- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Interaction of Adenovirus E1A with the HHV8 Promoter of Latent Genes: E1A Proteins are Able to Activate the HHV-8 LANAp in MV3 Reporter Cells
Karin Koehler-Hansner1, Ornella Flore2, Bertram Opalka 1, 4, Ulrich R Hengge*, 3
Abstract
Human herpesvirus 8 (HHV-8) is associated with Kaposi’s sarcoma, body cavity-based lymphoma, and Castleman’s disease. Adenoviral (Ad) E1A proteins regulate the activity of cellular and viral promoters/enhancers and transcription factors and can suppress tumorigenicity of human cancers. As (i) HHV-8 and Ad may co-exist in immunocompromised patients and (ii) E1A might be considered as therapeutic transgene for HHV-8-associated neoplasms we investigated whether the promoter of the latency-associated nuclear antigen (LANAp) controlling expression of vCyclin, vFLIP, and LANA proteins required for latent type infection is regulated by E1A. Transfection experiments in MV3 melanoma cells revealed activation of the LANAp by Ad5 E1A constructs containing an intact N terminus (aa 1-119). In particular, an Ad12 E1A mutant, Spm2, lacking six consecutive alanine residues in the “spacer” region activated the HHV-8 promoter about 15-fold compared to vector controls. In summary, we report the activation of the LANAp by E1A as a novel interaction of E1A with a viral promoter. These data may have relevance for the management of viral infections in immunocompromised patients. A role for E1A as a therapeutic in this context remains to be defined.
Article Information
Identifiers and Pagination:
Year: 2008Volume: 2
First Page: 61
Last Page: 68
Publisher Id: TOVJ-2-61
DOI: 10.2174/1874357900802010061
Article History:
Received Date: 30/5/2008Revision Received Date: 5/6/2008
Acceptance Date: 10/6/2008
Electronic publication date: 7/7/2008
Collection year: 2008
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/license/by/2.5/), which permits unrestrictive use, distribution, and reproduction in any medium, provided the original work is properly cited.
* Address correspondence to this author at the Department of Dermatology, Heinrich-Heine-University, Moorenstrasse 5, D-40225 Duesseldorf, Germany; Tel: +49-(0)211-811-8066; Fax: +49-(0)211-811-8830; E-mail: ulrich.hengge@uni-duesseldorf.de
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 30-5-2008 |
Original Manuscript | Interaction of Adenovirus E1A with the HHV8 Promoter of Latent Genes: E1A Proteins are Able to Activate the HHV-8 LANAp in MV3 Reporter Cells | |
INTRODUCTION
Human herpesvirus 8 (HHV-8) was discovered in Kaposi’s sarcoma (KS) biopsy samples [1Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma Science 1994; 266(5192): 1865-9.-3Hengge UR, Ruzicka T, Tyring SK, et al. Update on Kaposi's sarcoma and other HHV8 associated diseases. Part 2: pathogenesis, Castleman's disease, and pleural effusion lymphoma Lancet Infect Dis 2002; 2(6): 344-52.], primary effusion lymphomas (PEL) [4Fan W, Bubman D, Chadburn A, Harrington WJ, Cesarman E, Knowles DM. Distinct subsets of primary effusion lymphoma can be identified based on their cellular gene expression profile and viral association J Virol 2005; 79(2): 1244-51.], multicentric Castleman’s disease [5Gessain A, Sudaka A, Brière J, et al. Kaposi sarcoma-associated herpes-like virus (Human Herpesvirus Type 8) DNA sequences in multicentric Castleman´s disease: Is there any relevant association in non-human immunodeficiency virus-infected patients? Blood 199; 87(1): 414-6.], and plasmablastic lymphomas [6Dupin N, Diss TL, Kellam P, et al. HHV-8 is associated with a plasmablastic variant of Castleman disease that is linked to HHV-8-positive plasmablastic lymphoma Blood 2000; 95(4): 1406-2.] and has been linked to pathogenesis of these disorders. The HHV-8 genome consists of a double-stranded DNA of approximately 165 kb in size. Between conserved gene blocks, divergent regions contain unique viral proteins, some of which mimic cell cycle regulation and signal transduction proteins required for lytic replication and latency [7Russo JJ, Bohenzky RA, Chien MC, et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA 1996; 93(25): 14862-7.]. Highly variable proteins such as products of the K1 open-reading frame have been used for molecular epidemiology studies [8Kouri V, Liang X, Rodriguez ME, et al. Molecular epidemiology and KSHV K1 subtypes in Cuban AIDS-KS population AIDS 2005; 19(9): 984-7.]. In KS, HHV8 persists latently in the majority of tumor cells with fewer than 5% of cells undergoing spontaneous lytic replication [9Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus J Virol 1998; 72(10): 8309-15., 10Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. Transcription mapping of the Kaposi´s sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J Virol 1998; 72(2): 1005-2.]. Mapping of transcripts involved in latency pointed to responsible genes such as ORF73, ORF72 encoding vCyc, a functional viral cyclin, and ORFK13 encoding vFLIP, a protein [11Cesarman E, Knowles DM. Kaposi´s sarcoma-associated herpesvirus: a lymphotrophic human herpesvirus associated with Kaposi´s sarcoma, primary effusion lymphoma, and multicentric Castelman´s disease Sem Diagn Pathol 1997; 14(1): 54-66., 12Chang Y, Moore PS, Talbot SJ, et al. Cyclin encoded by KS herpesvirus Nature 1996; 382(6590): 410.] likely to have anti-apoptotic activity and contribute to the tumor phenotype [13Bertin J, Armstrong RC, Ottilie S, et al. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis Proc Natl Acad Sci USA 1997; 94(4): 1172-6.].
LANA, the latent nuclear antigen of HHV-8 (ORF73), is a multifunctional protein interacting with several cellular proteins suggesting a role in regulation of gene expression, and it has transforming activity in conjunction with H-ras [14Swaminathan S. Molecular biology of Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus Sem Hematol 2003; 40(2): 107-5.]. It tethers the viral episomal DNA to the host cell chromosomal DNA [15Cotter MA 2nd , Robertson ES . The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells Virology 1999; 264(1): 254-64.] and is able to bind p53, thus preventing apoptosis [16Friborg J Jr, Kong W, Hottiger MO, Nabel G. p53 inhibition by the LANA protein of KSHV protects against cell death Nature 1999; 402(6764): 889-94.]. Recent studies of HHV-8 latent replication and maintenance have revealed that two nuclear localization signals are located in the C-terminal region and N-terminal region of LANA [17Piolot T, Tramier M, Coppey M, Nicolas JC, Marechal V. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes J Virol 2001; 75(8): 3948-59.] whereas the C-terminal domain of LANA is required for dimerization and DNA binding [18Schwam DR, Luciano RL, Mahajan SS, Wong L, Wilson AC. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies J Virol 2000; 74(18): 8532-40.]. This interaction is necessary and sufficient for virus latency [19Garber AC, Hu J, Renne R. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication J Biol Chem 2002; 277(30): 27401-11.]. Suppression of the promoter of the latent genes may interrupt the latent life cycle of the virus and drive infected cells into apoptosis.
The E1A gene of human adenoviruses (Ad) is expressed early after infection and encodes multifunctional proteins that activate and suppress the transcription of both viral and cellular genes [20-22Gallimore PH, Turnell AS. Adenovirus E1A: remodelling the host cell, a life or death experience Oncogene 2001; 20(54): 7824-35.]. Due to alternative splicing Ad5 E1A encodes five different mRNAs. The two largest differentially spliced transcripts of Ad5 E1A encode polypeptides of 289 and 243 amino acids (aa) that account for most if not all of E1A’s biologic activities. Four conserved regions (CRs) CR1 to CR4 are highly homologous between different Ad serotypes, and the CR3 containing a transactivation domain resembles most of the 289aa protein-specific aa sequence [23Lillie JW, Green MR. Transcription activation by the adenovirus E1a protein Nature 1989; 338(6210): 39-44.]. The 243aa protein encodes all the functions necessary for immortalization of primary rodent cells or, in cooperation with other viral or cellular oncogenes, their full malignant transformation [24Graham FL, van der Eb AJ, Heijneker HL. Size and location of the transforming region in human adenovirus type 5 DNA Nature 1974; 251(5477): 687-91.-26Zerler B, Moran B, Maruyama K, Moomaw J, Grodzicker T, Ruley HE. Adenovirus E1A coding sequences that enable ras and pmt oncogenes to transform cultured primary cells Mol Cell Biol 1986; 6(3): 887-99.]. E1A proteins do not bind DNA themselves but control cell proliferation and transformation by modulating gene expression through interaction with several cellular proteins. These include p105RB or the transcriptional coactivator p300 that is involved in the regulation of many promoters. These activities of EIA have been reviewed extensively [22Gallimore PH, Turnell AS. Adenovirus E1A: remodelling the host cell, a life or death experience Oncogene 2001; 20(54): 7824-35., 27Brockmann D, Esche H. The multifunctional role of E1A in the transcriptional regulation of CREB/CBP-dependent target genes Curr Top Microbiol Immunol 2003; 272: 97-129.-36Whyte P, Ruley HE, Harlow E. Two regions of the adenovirus early region 1A proteins are required for transformation J Virol 1988; 62(1): 257-65.].
Despite their transforming potential in rodent cells E1A proteins have shown broad tumor suppressive activity in a variety of human tumor cells, including the suppression of metastatic potential and induction of susceptibility to cytostatic drugs and ionizing radiation [37Frisch SM. Antioncogenic effect of adenovirus E1A in human tumor cells Proc Natl Acad Sci USA 1991; 88(20): 9077-81.-39Frisch SM, Mymryk JS. Adenovirus-5 E1A. paradox and paradigm Nat Rev Mol Cell Biol 2002; 3(6): 441-52.]. In consequence, E1A has already found its way into clinical trials targeting HER2/neu-overexpressing ovarian and breast cancer as well as head and neck cancer [40Hortobagyi GN, Ueno NT, Xia W, et al. Cationic liposome-mediated E1A gene transfer to human breast and ovarian cancer cells and its biologic effects: a phase I clinical trial J Clin Oncol 2001; 19(14): 3422-3.-42Madhusudan S, Tamir A, Bates N, et al. A multicenter phase I gene therapy clinical trial involving intraperitoneal administration of E1A-lipid complex in patients with recurrent epithelial ovarian cancer overexpressing HER-2/neu oncogene Clin Cancer Res 2004; 10(9): 2986-96.]. Safety and feasibility of E1A gene therapy was established and in head and neck cancer patients a modest tumor response was observed [41Villaret D, Glisson B, Kenady D, et al. A multicenter phase II study of tgDCC-E1A for the intratumoral treatment of patients with recurrent head and neck squamous cell carcinoma Head Neck 2001; 24(7): 661-9.] warranting further evaluation of E1A as a therapeutic candidate drug.
Because E1A is known to regulate other viral promoters, both positively (e.g. CMV) [43Metcalf JP, Monick MM, Stinski MF, Hunninghake GW. Adenovirus E1A 13S gene product up-regulates the cytomegalovirus major immediate early promoter Am J Respir Cell Mol Biol 1994; 10(4): 448-52.] and negatively (e.g. the HIV-1 LTR) [44Song CZ, Loewenstein PM, Green M. Repression in vitro, by human adenovirus E1A protein domains, of basal or Tat-activated transcription of the human immunodeficiency virus type 1 long terminal repeat J Virol 1995; 69(5): 2907-11.], we asked whether E1A was also interfering with transcription from the LANAp of HHV-8. Since HHV-8 has been reported to latently infect a variety of adherent tumor cell lines of epithelial, endothelial, and mesenchymal origin [45Bechtel JT, Liang Y, Hvidding J, Ganem D. Host range of Kaposi's sarcoma-associated herpesvirus in cultured cells J Virol 2003; 77(11): 6474-81.], we used a reporter cell system throughout this study. In efforts to further improve the safety of E1A for clinical use we had found that in principle it is possible to separate transforming and reversing functions in a melanoma model [46Rumpf H, Esche H, Kirch HC. Two domains within the adenovirus Type 12 E1A unique spacer have disparate effects on the interaction of E1A with p105-Rb and the transformation of primary mouse cells Virology 1999; 257(1): 45-53., 47Dickopp A, Esche H, Swart G, Seeber S, Kirch HC, Opalka B. Transformation-defective adenovirus 5 E1A mutants exhibit antioncogenic properties in human BLM melanoma cells Cancer Gene Ther 2000; 7(7): 1043-50.] and thus included respective deletion mutants of E1A in the present study. Here we report on activation of the LANAp in E1A-transfected cells. This finding may - apart from it’s impact for a putative gene therapy with E1A - also be of interest in a more general way since infections with viruses are problematic in immunocompromised hosts, e.g. in settings of bone marrow or organ transplantation where co-infections with different viruses may occur [48Kusne S, Blair J. Viral and fungal infections after liver transplantation - part II Liver Transplant 2006; 12(1): 2-11.].
MATERIALS AND METHODOLOGY
Cells
MV3, a human melanoma cell line [49van Muijen GN, Jansen KF, Cornelissen IM, Smeets DF, Beck JL, Ruiter DJ. Establishment and characterization of a human melanoma cell line (MV3) which is highly metastatic in nude mice Int J Cancer 1991; 48(1): 85-91.] and BC-3 cells, a human PEL-derived B-cell line coinfected with HHV-8 were cultured as described [50Arvanitakis L, Mesri EA, Nador RG, et al. Establishment and characterization of a primary (body cavity-based) lymphoma cell line (BC-3) harbouring kaposi´s sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus Blood 1996; 88(7): 2648-54., 51Kirch HC, Ruschen D, Brockmann D, et al. Tumor-specific activation of hTERT-derived promoters by tumor suppressive E1A-mutants involves recruitment of p300/CBP/HAT and suppression of HDAC-1 and defines a combined tumor targeting and suppression system Oncogene 2001; 21(52): 7991-8000.].
Plasmids
Construction of the luciferase gene plasmids HHV-8 WT promoter, del -262 and del -67 was described previously [52Sarid R, Wiezorek JS, Moore PS, Chang Y. Characterization and cell cycle regulation of the major Kaposi´s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter J Virol 1999; 73(2): 1438-46.]. A del-59 reporter gene plasmid was constructed on the basis of the del-67 construct as follows: The LP1/2 fragment upstream from nt +30 relative to the start site was synthesized by PCR with an antisense primer (+9 to +30) 5´-CCG CTCGAGCTGCCTCCAAATGATACACA-3´ and sense primer 88 bp HHV-8 (5´-CGGGGTACCCCAATCAGAAA GTAGCTTGA-3´) with the reporter gene plasmid 96 bp HHV-8 (pGL3.1) as template [52Sarid R, Wiezorek JS, Moore PS, Chang Y. Characterization and cell cycle regulation of the major Kaposi´s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter J Virol 1999; 73(2): 1438-46.]. A second reporter gene plasmid, del-59Sp-1, was constructed using the Quick-Change Multi Site-Directed Mutagenesis Kit (Stratagene, Heidelberg). The Sp-1 binding site was deleted according to the instructions of the manufacturer using the two primers 5´-GTCCCGGGCGCCGCGTAGTGGAAGTTATATCAAG CTACTTTCTGATTG-3´ (sense) and 5´-CAATCAGAAAG TAGCTTGATATAACTTCCACTACGCGGCGCCCGGGA C-3´ (antisense) with the reporter gene plasmid and the 88 bp HHV-8 as templates. The E1A mutants used in this study were described previously [46Rumpf H, Esche H, Kirch HC. Two domains within the adenovirus Type 12 E1A unique spacer have disparate effects on the interaction of E1A with p105-Rb and the transformation of primary mouse cells Virology 1999; 257(1): 45-53., 47Dickopp A, Esche H, Swart G, Seeber S, Kirch HC, Opalka B. Transformation-defective adenovirus 5 E1A mutants exhibit antioncogenic properties in human BLM melanoma cells Cancer Gene Ther 2000; 7(7): 1043-50., 51Kirch HC, Ruschen D, Brockmann D, et al. Tumor-specific activation of hTERT-derived promoters by tumor suppressive E1A-mutants involves recruitment of p300/CBP/HAT and suppression of HDAC-1 and defines a combined tumor targeting and suppression system Oncogene 2001; 21(52): 7991-8000.]. The Spm2 construct expressed the Spm2 E1A mutant fused to the herpes virus VP22 tegument protein to increase intercellular spread [53Elliot G, O´Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein Cell 1997; 88(2): 223-33.].
Transient Transfections
MV3 cells were transiently transfected with JetPEI (Qbiogene, Heidelberg, Germany) in 24-well plates. Transfection mixes contained 1 µg of a HHV-8 promoter luciferase reporter plasmid and 1 ng, 10 ng, or 50 ng of E1A expression plasmids. Cells were lysed and luciferase activity was measured. Transfections of BC-3 cells were performed by electroporation (Bio-Rad Gene Pulser II, Bio-Rad Laboratories, Munich, Germany) at settings of 230 mV and 975 µF in 0.4 cm cuvettes (Bio-Rad), using 2.9 x 107 cells/cuvette resuspended in 0.35 ml RPMI 1640 medium supplemented with 15 % FCS and 1 % glutamine. Luciferase reporter constructs were constructed with the promoter of the latent genes of HHV-8 and its deletion mutants being fused to the luciferase vector pGL3b (Promega, Mannheim, Germany). The positive control plasmid pGL3c was obtained from Promega, the negative control plasmids pRc/RSV and pVP22/myc-His used in this study were purchased from Invitrogen. Transfection experiments were performed three times and in triplicate. Transfections/electroporations with the pGL3c plasmid expressing the luciferase gene from the SV40 promoter/enhancer or a human telomerase reverse transcriptase (hTERT) promoter were included to score for successful transduction of the cells (data not shown).
Luciferase Assays
Harvested cells were washed once with 1 ml PBS and lysed in 200 µl of 1 x Glo Lysis buffer (Promega) for the cells transfected with JetPEI. Luciferase activity was measured according to the manufacturer´s recommendations (Promega, Mannheim, Germany). Results are given as relative luciferase activity or fold induction compared to the negative control plasmids, i.e. pRc/RSV or pVP22/myc-His for the experiments with Spm2 E1A; mean values +/- SD are depicted.
Statistical Analysis
For statistical analysis the student’s t-test was used throughout all investigations.
RESULTS
Basal Activity of the LANAp in MV3 Cells
To confirm that the LANAp operates in MV3 melanoma cells, we assessed its ability to drive expression of a reporter gene in transiently transfected MV3 cells. Promoter-reporter gene recombinants were used where LANAp stretches of various length (Fig. 1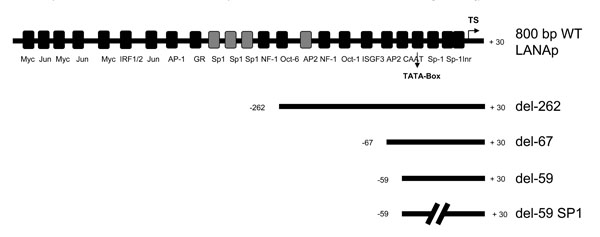 ) were cloned upstream to the reporter gene cassette.
) were cloned upstream to the reporter gene cassette.
 |
Fig. (1) Schematic representation of the LANAp (not drawn to scale) and deletion mutants used in our studies. Transcription factor binding sites are also indicated. |
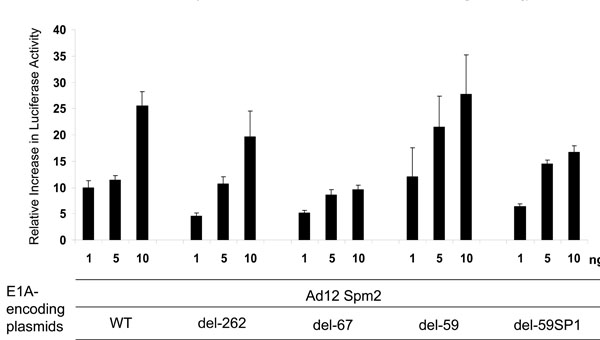 |
Fig. (5) Comparison of Ad12 Spm2 E1A-mediated dose-dependent activation of transcription in MV3 cells between the del 59 SP1, the del -59, the del -67, the del -262 and the complete 804 bp HHV-8 promoter constructs. The transfection procedure was as described in Fig. (4 |
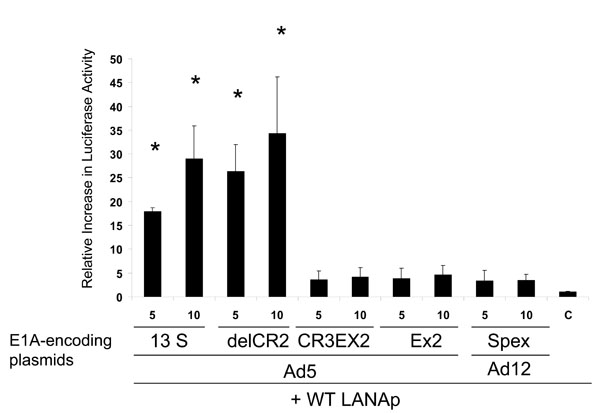 |
Fig. (6) The HHV-8 promoter is transactivated by Ad5 E1A and the E1A deletion mutant delCR2. E1A derivatives lacking an intact aminoterminus were not able to transactivate the LANAp. The transfection procedure was as described in Fig. (4 |
The fusion sites were located in the 5´ untranslated leader sequence of the LT1 / 2 transcripts corresponding to nt 127-870 [52Sarid R, Wiezorek JS, Moore PS, Chang Y. Characterization and cell cycle regulation of the major Kaposi´s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter J Virol 1999; 73(2): 1438-46.]. Results from three independent experiments performed in triplicate are shown in Fig. (2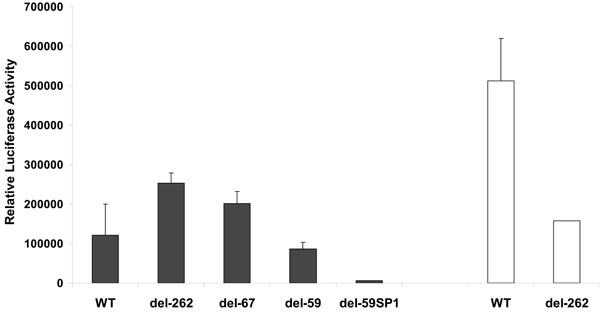 ). When transfected into MV3 cells, an 804 bp HHV-8 WT promoter construct containing several transcription factor binding sites stimulated luciferase expression. Truncation of the WT LANAp from the 5´ end enhanced the basal promoter activity, and a construct containing only 67 bp was able to initiate reporter transcription comparable to the del-262 plasmid. Further deletion of the promoter caused a decline in basal activity (Fig. 2
). When transfected into MV3 cells, an 804 bp HHV-8 WT promoter construct containing several transcription factor binding sites stimulated luciferase expression. Truncation of the WT LANAp from the 5´ end enhanced the basal promoter activity, and a construct containing only 67 bp was able to initiate reporter transcription comparable to the del-262 plasmid. Further deletion of the promoter caused a decline in basal activity (Fig. 2 ). The del-59 HHV-8 promoter even showed a lower basal activity than the WT LANAp. In the HHV-8-positive B-cell line BC3 higher luciferase activity was observed under the control of the WT and the -262 LANAp (Fig. 2
). The del-59 HHV-8 promoter even showed a lower basal activity than the WT LANAp. In the HHV-8-positive B-cell line BC3 higher luciferase activity was observed under the control of the WT and the -262 LANAp (Fig. 2 ). Taken together, these experiments confirmed transcriptional activity of the LANAp in MV3 melanoma cells that were used in further cotransfection experiments.
). Taken together, these experiments confirmed transcriptional activity of the LANAp in MV3 melanoma cells that were used in further cotransfection experiments.
E1A Transactivates the LANAp
In order to examine transformation-negative Ad2 or Ad5 and Ad12 E1A constructs for use as tumor-suppressive transgenes in gene therapy of malignant disease we had cloned a series of plasmids encoding the WT 13S and 12S mRNAs and various truncated forms thereof (Fig. 3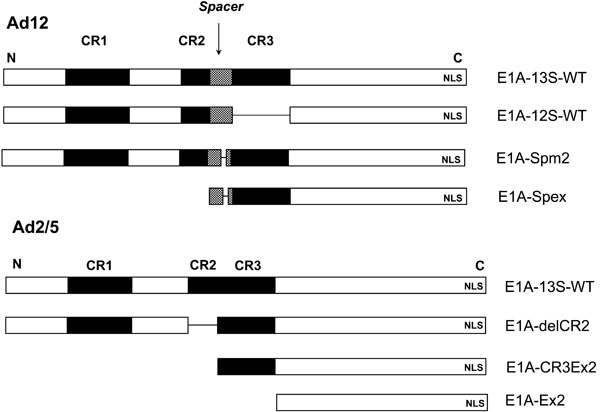 ) [47Dickopp A, Esche H, Swart G, Seeber S, Kirch HC, Opalka B. Transformation-defective adenovirus 5 E1A mutants exhibit antioncogenic properties in human BLM melanoma cells Cancer Gene Ther 2000; 7(7): 1043-50., 51Kirch HC, Ruschen D, Brockmann D, et al. Tumor-specific activation of hTERT-derived promoters by tumor suppressive E1A-mutants involves recruitment of p300/CBP/HAT and suppression of HDAC-1 and defines a combined tumor targeting and suppression system Oncogene 2001; 21(52): 7991-8000.].
) [47Dickopp A, Esche H, Swart G, Seeber S, Kirch HC, Opalka B. Transformation-defective adenovirus 5 E1A mutants exhibit antioncogenic properties in human BLM melanoma cells Cancer Gene Ther 2000; 7(7): 1043-50., 51Kirch HC, Ruschen D, Brockmann D, et al. Tumor-specific activation of hTERT-derived promoters by tumor suppressive E1A-mutants involves recruitment of p300/CBP/HAT and suppression of HDAC-1 and defines a combined tumor targeting and suppression system Oncogene 2001; 21(52): 7991-8000.].
We here examined whether transcription from the LANAp was modulated by Ad2 and Ad12 E1A constructs in MV3 reporter cells (Fig. 4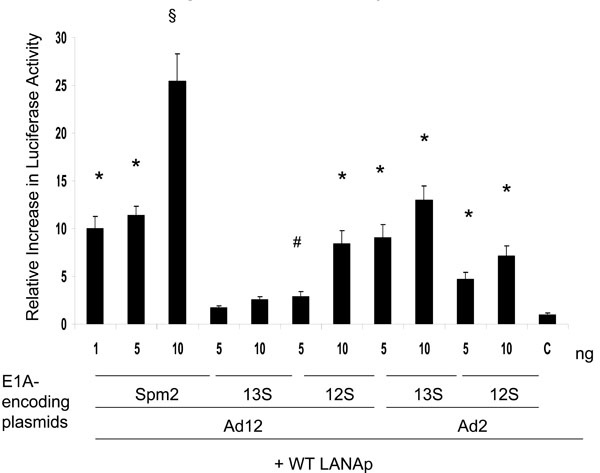 ). We found a 5 to 13-fold activation of transcription from the WT LANAp induced by Ad2 E1A while the VP22 control plasmid showed no transactivation (Fig. 4
). We found a 5 to 13-fold activation of transcription from the WT LANAp induced by Ad2 E1A while the VP22 control plasmid showed no transactivation (Fig. 4 ).
).
Ad12 12S E1A caused a dose-dependent transactivation of the LANAp. In particular, we observed significant transactivation capacity (up to 25-fold) after transfection of the Ad12 Spm2 E1A plasmid (Fig. 5 ). The E1A-derivative Spm2 used in these experiments was fused to the VP22 HSV tegument protein [51Kirch HC, Ruschen D, Brockmann D, et al. Tumor-specific activation of hTERT-derived promoters by tumor suppressive E1A-mutants involves recruitment of p300/CBP/HAT and suppression of HDAC-1 and defines a combined tumor targeting and suppression system Oncogene 2001; 21(52): 7991-8000.] which mediates intercellular transport of the fusion protein [53Elliot G, O´Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein Cell 1997; 88(2): 223-33.] and may thus allow a more efficient delivery of the transgene product. Furthermore, E1A-Spm2 was able to enhance the transactivation from the WT LANAp as well as from its deletion mutants -262, -67, -59, and even -59SP1. The latter construct lacks one of the most proximal SP1 sites (Fig. 5
). The E1A-derivative Spm2 used in these experiments was fused to the VP22 HSV tegument protein [51Kirch HC, Ruschen D, Brockmann D, et al. Tumor-specific activation of hTERT-derived promoters by tumor suppressive E1A-mutants involves recruitment of p300/CBP/HAT and suppression of HDAC-1 and defines a combined tumor targeting and suppression system Oncogene 2001; 21(52): 7991-8000.] which mediates intercellular transport of the fusion protein [53Elliot G, O´Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein Cell 1997; 88(2): 223-33.] and may thus allow a more efficient delivery of the transgene product. Furthermore, E1A-Spm2 was able to enhance the transactivation from the WT LANAp as well as from its deletion mutants -262, -67, -59, and even -59SP1. The latter construct lacks one of the most proximal SP1 sites (Fig. 5 ) and was silent in the MV3 cells (Fig. 2
) and was silent in the MV3 cells (Fig. 2 ).
).
Activation of Transcription from the LANAp by E1A Requires Aminoterminal E1A Sequences
To further investigate functional domains of the E1A proteins involved in transactivation of the LANAp, we studied whether deletion mutants of E1A were still able to activate this promoter. We found a 18 to 35-fold activation of transcription from the LANAp in the MV3 melanoma cell line by those Ad5 E1A constructs that contained an intact aminoterminus (13S and delCR2, Fig. (6 ); transfection efficiency: 50 ± 15% each). The Ad5 E1A-CR3Ex2, Ad12 E1A-Spex and Ad5 E1A-Ex2 constructs had essentially no effect.
); transfection efficiency: 50 ± 15% each). The Ad5 E1A-CR3Ex2, Ad12 E1A-Spex and Ad5 E1A-Ex2 constructs had essentially no effect.
DISCUSSION
In this study we report evidence that the LANAp of HHV-8 can be up-regulated by Ad2 or Ad5 and Ad12 E1A gene products in MV3 human melanoma reporter cells. The MV3 reporter cell line was chosen for this initial series of experiments because it was easy to transfect. Moreover, studies to define regulatory elements of the LANAp and other latency-associated HHV-8 genes have been previously performed by others in a variety of adherent, non B-lymphoid cell types, including HeLa, HEK293, and SLK cells demonstrating the general usability of these systems [52Sarid R, Wiezorek JS, Moore PS, Chang Y. Characterization and cell cycle regulation of the major Kaposi´s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter J Virol 1999; 73(2): 1438-46., 54Jeong J, Papin J, Dittmer D. Differential regulation of the overlapping Kaposi´s Sarcoma-Associated Herpesvirus vGCR (orf74) and LANA (orf73) promoters J Virol 2001; 75(4): 1798-807., 55Jeong JH, Orvis J, Kim JW, McMurtrey CP, Renne R, Dittmer DP. Regulation and Auto-regulation of the promoter for the latency-associated nuclear antigen (LANA) of Kaposi´s sarcoma-associated herpesvirus J Biol Chem 2004; 279(16): 16822-31.]. As shown here, the WT LANAp had basal activity in the MV3 cells, although to a lesser extent than in B lymphoid BC-3 cells.
We detected an increase in basal LANAp activity upon truncation with the highest activity being confined to the -262 construct and the lowest confined to pos. -67. This is inconsistent with the data from others who described a low activity of LANAp deletion constructs in HeLa cells and only minor influences of LANAp truncation down to -67 in BJAB cells [52Sarid R, Wiezorek JS, Moore PS, Chang Y. Characterization and cell cycle regulation of the major Kaposi´s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter J Virol 1999; 73(2): 1438-46.]. The discrepancy between our data and the findings of Sarid et al. may be due to different cell lines since we found a 14-fold lower basal activity of the WT promoter in HeLa cells in comparison with MV3 cells (data not shown). Deletion of one of the postulated two SP-1 sites on the del-59 construct reduced the activity of the LANAp by about 90% suggesting that the first SP1 binding site downstream of the TATA box at position -27 is essential for LANAp promoter activity. We further suggest that the AP-2α binding site upstream of the TATA box in addition to the SP1 binding sites downstream of the TATA box is necessary for efficient basal activation. Taken together, these data establish positive regulatory elements in the regions downstream of pos -262 and between pos. -67 and -1 of the LANAp, respectively, at least for its regulation in MV3 cells. The LANAp region spanning pos. -263 to -804 harbors single or multiple binding sites for at least Sp-1, GR, Ap-1, Irf1/2, Myc, and Jun [52Sarid R, Wiezorek JS, Moore PS, Chang Y. Characterization and cell cycle regulation of the major Kaposi´s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter J Virol 1999; 73(2): 1438-46.]. The binding sites responsible for the suppression observed in multiple cell systems remain to be defined; however, sequence analysis software (www. gene-regulation.com/cgi-bin/pub/programs/alibaba2/webbaba 2.cgi) identified a suppressor element at pos. -440 of the LANAp (Köhler, unpublished data). It should be noted that this negative regulation was not observed in BC-3 cells, consistent with findings of Jeong et al. in BJAB cells and BC-3 cells of lymphocytic origin [54Jeong J, Papin J, Dittmer D. Differential regulation of the overlapping Kaposi´s Sarcoma-Associated Herpesvirus vGCR (orf74) and LANA (orf73) promoters J Virol 2001; 75(4): 1798-807.]. Of note, Jeong et al. observed a substantial increase of luciferase activity driven by LANAp constructs in adherent cells like HEK293 when sequences from pos. -1490 to -279 were deleted [54Jeong J, Papin J, Dittmer D. Differential regulation of the overlapping Kaposi´s Sarcoma-Associated Herpesvirus vGCR (orf74) and LANA (orf73) promoters J Virol 2001; 75(4): 1798-807.]. In summary, we showed that a minimal region starting at position -67 from the transcription start site defined at nt 127-900 [52Sarid R, Wiezorek JS, Moore PS, Chang Y. Characterization and cell cycle regulation of the major Kaposi´s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter J Virol 1999; 73(2): 1438-46.] or 127-880 [55Jeong JH, Orvis J, Kim JW, McMurtrey CP, Renne R, Dittmer DP. Regulation and Auto-regulation of the promoter for the latency-associated nuclear antigen (LANA) of Kaposi´s sarcoma-associated herpesvirus J Biol Chem 2004; 279(16): 16822-31.] is necessary and sufficient for constitutive promoter activity and also for robust expression from the LANAp in the MV3 cell line.
When low amounts of plasmids encoding the 13S and 12S cDNAs of Ad2 and Ad12, respectively, were cotransfected with the LANAp constructs, a dose-dependent activation of the WT LANAp was detected in MV3 cells. A more pronounced induction was found when the Ad12 Spm2 mutant was used. It is of particular interest that Spm2 induced transcription from truncated LANAps down to the del-59 construct and even from the del-59Sp-1 mutant. Thus, Spm2 E1A seems to be able to override (i) the influence of the negative regulatory element between pos. -263 and -804 in MV3 cells and, moreover, (ii) the requirement for the Sp-1-site that is deleted in the construct del-59Sp-1 for basal LANAp activity in these cells. Spm2, a transformation-defective derivative of Ad12 E1A lacking six consecutive alanines in the Ad12-specific so-called “spacer” region impairing binding to Rb [46Rumpf H, Esche H, Kirch HC. Two domains within the adenovirus Type 12 E1A unique spacer have disparate effects on the interaction of E1A with p105-Rb and the transformation of primary mouse cells Virology 1999; 257(1): 45-53.] is able to stimulate transcription from the human TERT promoter in melanoma cells without activating TERT in non-transformed fibroblasts [51Kirch HC, Ruschen D, Brockmann D, et al. Tumor-specific activation of hTERT-derived promoters by tumor suppressive E1A-mutants involves recruitment of p300/CBP/HAT and suppression of HDAC-1 and defines a combined tumor targeting and suppression system Oncogene 2001; 21(52): 7991-8000.]. Moreover, there is some evidence that Spm2 may harbor tumor-suppressive activity in vivo (Kirch et al., unpublished observations). However, the Spm2 construct contained the VP22 domain considered to enhance cell-to-cell spread of fusion proteins [56Zender L, Kuhnel F, Kock R, Manns M, Kubicka S. VP22-mediated intercellular transport of p53 in hepatoma cells in vitro and in vivo Cancer Gene Ther 2002; 9(6): 489-96.], thereby possibly increasing the protein dosage per cell as well as the number of cells transduced. As E1A has recently been reported to have dramatically different effects depending on expression levels in the 3T3L1 preadipocytic cell line [57Cao J, Arulanandam R, Vultur A, et al. Adenovirus-5 E1A suppresses differentiation of 3T3 L1 preadipocytes at lower levels than required for induction of apoptosis Mol Carcinogen 2005; 43(1): 38-50.], such gene dosage effects have to be seriously considered and await further investigation. We and others have previously ruled out a direct transactivating effect of VP22 sequences [47Dickopp A, Esche H, Swart G, Seeber S, Kirch HC, Opalka B. Transformation-defective adenovirus 5 E1A mutants exhibit antioncogenic properties in human BLM melanoma cells Cancer Gene Ther 2000; 7(7): 1043-50., 51Kirch HC, Ruschen D, Brockmann D, et al. Tumor-specific activation of hTERT-derived promoters by tumor suppressive E1A-mutants involves recruitment of p300/CBP/HAT and suppression of HDAC-1 and defines a combined tumor targeting and suppression system Oncogene 2001; 21(52): 7991-8000., 56Zender L, Kuhnel F, Kock R, Manns M, Kubicka S. VP22-mediated intercellular transport of p53 in hepatoma cells in vitro and in vivo Cancer Gene Ther 2002; 9(6): 489-96.].
N-terminal sequences of E1A, i.e. aa positions 1-119, proved to be a necessary prerequisite for activation of the LANAp. This region of E1A contains transcriptional activation properties in which binding sites for a multitude of transcriptional activators and enhancers are located, in particular AP-2- and CBP/p300-binding sites [27Brockmann D, Esche H. The multifunctional role of E1A in the transcriptional regulation of CREB/CBP-dependent target genes Curr Top Microbiol Immunol 2003; 272: 97-129.].
In contrast to others [54Jeong J, Papin J, Dittmer D. Differential regulation of the overlapping Kaposi´s Sarcoma-Associated Herpesvirus vGCR (orf74) and LANA (orf73) promoters J Virol 2001; 75(4): 1798-807., 55Jeong JH, Orvis J, Kim JW, McMurtrey CP, Renne R, Dittmer DP. Regulation and Auto-regulation of the promoter for the latency-associated nuclear antigen (LANA) of Kaposi´s sarcoma-associated herpesvirus J Biol Chem 2004; 279(16): 16822-31.] we did not detect any autoactivation of the LANAp by cotransfection of plasmids encoding LANA itself or a combination of vFLIP, vCyclin, and LANA (data not shown). Moreover, cotransfection of these plasmids did not interfere with stimulation of transcription from the LANAp by Spm2 E1A. Mechanisms for these observations remain to be elucidated.
In a preliminary series of experiments neither cotransfection of Ad2 E1A nor Ad12 E1A Spm2 with the WT LANAp in HHV-8-positive BC-3 cells resulted in any substantial activation of the LANAp (data not shown). Whether these observations reflect a biological phenomenon or rather technical problems remains unclear; in the BC-3 cells we also observed only a marginal activation of a Hsp70 positive control promoter that was highly active in the MV3 cells (data not shown). The RSV promoter controlling E1A expression in most of our constructs has been reported to be active in B-lymphoid cells [58Zarrin AA, Malkin L, Fong I, Luk KD, Ghose A, Berinstein NL. Comparison of CMV, RSV, SV40 viral and Vlambda1 cellular promoters in B and T lymphoid and non-lymphoid cell lines Biochim Biophys Acta 1999; 1446(1-2): 135-9.]. Pilot experiments using a GFP expression plasmid revealed a transfer efficiency of 55±15 % when the optimized electroporation protocol was used and 40 ± 12.5% as scored by fluorescence microscopy when cotransfections were performed (data not shown). Further experimental efforts will be required to test whether our findings reported here hold also true for the context of a B cell environment.
Recently, novel regulatory elements and mechanisms controlling protein expression at the RNA level have been described. These are miRNAs that can induce specific silencing of mRNAs and the RNA editing, the post-transcriptional sequence modification of primary transcripts. MiRNAs are also encoded by viruses and can be orthologous to cellular counterparts [59Gottwein E, Mukherjeee N, Sachse C, et al. A viral microRNA functions as an orthologue of cellular mir-155 Nature 2007; 450(7172): 1096-9.]. A cluster encoding HHV-8-specific miRNAs has been found within a region transcribed under the control of the LANAp as well as other promoters [60Pearve M, Matsumura S, Wilson AC. Transcripts encoding K12, v-FLIP, v-Cyclin, and the microRNA cluster of Kaposi's sarcoma-associated herpesvirus originate from a common promoter J Virol 2005; 79(22): 14457-64.]. RNA editing [61Linnstaedt SD, Kasprzak WK, Shapiro BA, Casey JL. The role of a metastable RNA secondary structure in hepatitis delta virus genotype III RNA editing RNA 2005; 12(8): 1521-33.] has been shown to control the biological functions of the HHV-8 Kaposin protein [62Gandy SZ, Linnstaedt SD, Mauralidhar S, Cashman KA, Rosenthal LJ, Casey JL. RNA editing of the human herpesvirus 8 kaposin transcript eliminates its transforming activity and is induced during lytic replication J Virol 2007; 81(24): 13544-51.]. These results - only very briefly mentioned here – call for the need to further elucidate the more complex regulation of protein expression by tumor-inducing viruses like HHV-8.
CONCLUSIONS
Taken together, we show, to our knowledge for the first time, that E1A can regulate and activate the LANAp of HHV-8, at least in MV-3 reporter cells. Thus, our data add the LANAp to the many cellular and viral promoters modulated by E1A. Whether this holds true for naturally HHV-8-containing cells remains to be determined since technical problems with our experiments failing to prove this in BC-3 cells cannot be ruled out at present. Whether this activity of E1A has implications for its use as a gene therapeutic remains elusive. The observation that downregulation of LANA by the small molecular weight component glycyrrhizic acid can induce apoptosis [63Curreli F, Friedman-Kien AE, Flore O. Glycyrrhizic acid alters Kaposi sarcoma-associated herpesvirus latency, triggering p53-mediated apoptosis in transformed B lymphocytes J Clin Invest 2005; 115(3): 642-52.] would favor a downregulation by E1A, too. In this respect our data would caution the therapeutic application of E1A in HHV-8-positive tumor patients. However, it is not yet known whether a maintenance of HHV-8 latency by E1A-mediated upregulated LANA rather than conversion to the lytic cycle might be beneficial, maybe in combination with cytotoxic or antiviral drugs. For severely immunocompromised patients, e.g. after organ transplantation, possibly infected among other viruses by HHV-8 and Ad concomitantly it may be of avail to know that Ad’s gene products can interact with HHV-8 regulatory elements, presumably in an undesired fashion.
ACKNOWLEDGEMENTS
We would like to thank Dr. H. C. Kirch for helpful advice and discussions, K. Thiel for expert technical assistance, Dr. W. Bardenheuer for technical support, and Dr. Y. Chang for the kind gift of reagents. This work has been supported by the Joachim Kuhlmann AIDS Foundation, Essen, Germany.





