- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Highly Sensitive Detection and Genotyping of HPV by PCR Multiplex and Luminex Technology in a Cohort of Colombian Women with Abnormal Cytology
Dabeiba A García1, Angel Cid-Arregui*, 2, Markus Schmitt3, Marcos Castillo4, 5, Ignacio Briceño4, Fabio A. Aristizábal66
Abstract
Cancer of the uterine cervix (CC) is the second most common cancer in women worldwide. In Colombia, CC is the second most frequent cancer among the entire women population and the first among women aged between 15 and 44 years, with an estimated incidence of 24.9 cases/100,000 inhabitants. The main risk factor is infection with one or more high-risk human papillomavirus (HPV) types. The aim of this study was to estimate the genotype-specific prevalence of human papillomavirus (HPV) DNA in patients with cervical pathology using the multiplex PCR and Luminex xMAP technology. In addition, we compared genotyping with Luminex xMAP and with Reverse Line Blot (RLB). A cohort of 160 patients participated in the study, of which 25.6% had no cervical lesions, 35% presented cervical intraepithelial neoplasia of grade I (CIN I), 10% CIN II, 20.6% CIN III and 8.8% CC. The most frequent viral types in all lesion grades were HPV16 and HPV18. Infections by a unique virus were less frequent (19.4%) than multiple infections (80.6%). Single infections were found in 22% of women with no cervical lesions, and in 14.3% of CIN I, 18.7% CIN II, 21.2% CIN III and 28.6% of CC. Multiple infections were observed in 78.0% of cervical samples with negative histopathologic diagnosis, and in 85.7% of CIN I, 81.2% CIN II, 78.8% CIN III and 71.4% CC. All samples analyzed with Luminex xMAP were HPV-positive, while we could detect HPV in only 48.8% of cases with RLB. Of the samples positive by both methods, there was a 67.2% correlation in the viral type(s) detected. In conclusion, Luminex suspension array showed a remarkably higher sensitivity compared with RLB. Multiple infections were unexpectedly common, being HPV types 16 and 18 the most prevalent in all histopathologic grades.
Article Information
Identifiers and Pagination:
Year: 2011Volume: 5
First Page: 70
Last Page: 79
Publisher Id: TOVJ-5-70
DOI: 10.2174/1874357901105010070
Article History:
Received Date: 11/2/2011Revision Received Date: 12/5/2011
Acceptance Date: 16/5/2011
Electronic publication date: 17/6/2011
Collection year: 2011
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Translational Immunology Group D015, German Cancer Research Center (DKFZ), Im Neuenheimer Feld 280, D-69120 Heidelberg, Germany; Tel: +49 6221 567109; Fax: +49 6221 564776; E-mail: a.cid@dkfz.de
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 11-2-2011 |
Original Manuscript | Highly Sensitive Detection and Genotyping of HPV by PCR Multiplex and Luminex Technology in a Cohort of Colombian Women with Abnormal Cytology | |
INTRODUCTION
Cancer of the cervix (CC) is the second most common cancer in women worldwide, with estimated 500,000 new cases of CC per year. In developing countries CC remains as a major cause of morbidity and mortality in women [1Saleem A, Tristram A, Fiander A, Hibbitts S. Prophylactic HPV vaccination: a major breakthrough in the fight against cervical cancer? Minerva Med 2009; 100: 503-23., 2Saslow D, Castle PE, Cox JT, et al. American Cancer Society Guideline for Human Papillomavirus (HPV) vaccine use to prevent cervical cancer and its precursors CA Cancer J Clin 2007; 57: 7-28.]. However, CC is a highly curable disease if detected at early stages [3World Health Organization. State of the art of new vaccines research and development. Initiative for Vaccine Research In: Geneva (Switzerland): World Health Organization 2006; pp. 96-8., 4Thompson J, Thomas LK, Shroyer KR. Human papillomavirus: molecular and cytologic/histologic aspects related to cervical intraepithelial neoplasia and carcinoma Hum Pathol 2008; 9: 154-66.]. Early detection is effective because the precursor lesions develop slowly preceding invasive cancer, typically over a period greater than 10 years [5Unger ER, Barr E. Human papillomavirus and cervical cancer Emerg Infect Dis 2004; 10: 2031-.]. These precursor lesions, the so-called cervical intraepithelial neoplasia (CIN) grades I to III, are detectable by cervical cytology. In countries where cytology has been introduced, CC rates have declined substantially [5Unger ER, Barr E. Human papillomavirus and cervical cancer Emerg Infect Dis 2004; 10: 2031-.]. A subset of human Papillomavirus (HPV) types plays an essential role in CC development [6Kisseljov FL, Kisseljova NP, Kobzeva VK, et al. State of human papilloma virus DNA in cervical carcinomas Mol Biol 2001; 35: 399-404., 7Cid-Arregui A. Therapeutic vaccines against human papillomavirus and cervical cancer Open Virol J 2009; 3: 67-83.]. Infection by oncogenic HPV types is considered a prerequisite for the development of this neoplasm, and about 99% of cervical cancers are HPV-infected [8Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide J Pathol 1999; 189: 12-9., 9Bosch FX, de San Jose S. Human papillomavirus and cervical cancer: burden and assessment of causality J Natl Cancer Inst Monogr 2003; 31: 3-13.] although most infections are cleared by the immune system [5Unger ER, Barr E. Human papillomavirus and cervical cancer Emerg Infect Dis 2004; 10: 2031-.]. However, infection with HPV per se is not sufficient for cancer to develop, since a number of risk co-factors have been identified, such as early initiation of sexual intercourse, higher number of sexual partners, concurrent viral and bacterial infections, use of oral contraceptives, dietary habits, multiparity and smoking, among others [4Thompson J, Thomas LK, Shroyer KR. Human papillomavirus: molecular and cytologic/histologic aspects related to cervical intraepithelial neoplasia and carcinoma Hum Pathol 2008; 9: 154-66., 10Reeves WC, Brinton LA, García M, et al. Human papillomarivus infection and cervical cancer in Latin America N Engl J Med 1989; 320: 1437-41.].
Approximately 118 HPV types have been fully characterized to date. Of these, a subset infect the mucosa of the genital tract and have been divided into three groups: 1) high-risk (HR-HPV), including types 16, 18, the most prevalent in CC; 2) possibly high-risk (pHR-HPV); and 3) low-risk (LR-HPV) HPV [11Schmitt M, Dondog B, Waterboer T, Pawlita M. Homogeneous amplification of genital human alpha papillomaviruses by PCR using novel broad-spectrum GP5+ and GP6+ primers J Clin Microbiol 2008; 46: 1050-9.].
Because just high-risk HPVs are etiologically linked to CC, efforts have been made to improve the sensitivity of the methods for virus type detection in samples from women with abnormal genital cytology. HPV detection and typing has become an essential tool for diagnostics and management of HPV-related disease. Molecular biology techniques, particularly PCR, made possible highly sensitive methods for detection of HPV DNA in cervical samples [12Gravitt P, Hakenewerth A, Stoerker J. A direct comparison of methods proposed for use in widespread screening of human papillomavirus infection Mol Cell Probes 1991; 5: 65-72.-14Cope JU, Hildesheim A, Schiffman MH, et al. Comparison of the hybrid capture tube test and PCR for detection of human papillomavirus DNA in cervical specimens J Clin Microbiol 1997; 35: 2262-5.]. HPV genotype can be determined analyzing the PCR products by hybridization with type-specific probes, sequencing or restriction fragment length polymorphism. One of the most useful PCR methods for genotyping HPV has been nonradioactive reverse line blot (RLB), which uses the general primers GP5+/GP6+ (GP) to amplify the L1 region of 37 mucosotropic HPV types [15Van Den Brule A, Pol R, Fransen-Daalmeijer N, Schouls L, Meijer C, Snijders P. GP5+/GP6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes J Clin Microbiol 2002; 40: 779-87.]. The technique is based on hybridization of oligonucleotide probes specific for each viral type with biotinylated PCR products obtained from clinical specimens, which are detected with streptavidin-peroxidase, revealed with enhanced chemiluminiscence (ECL), and finally exposed to an autoradiography film. The RLB system has proven most useful for typing of samples that were prescreened by HPV group-specific assays, such as HR-HPV or LR-HPV GP5+/6+ PCR-EIA or Hybrid Capture 2.
In recent years a multiplex HPV genotyping assay was developed based on GP PCR followed by subsequent detection of the biotinylated products with 22 type-specific oligonucleotide probes covalently coupled to distinct sets of fluorescently-labeled polystyrene beads [16Schmitt M, Bravo I, Snijders P, Gissmann L, Pawlita M, Waterboer T. Bead-Based Multiplex Genotyping of Human Papillomaviruses J Clin Microbiol 2006; 44: 504-12.]. (Luminex Technology). This method uses a broad spectrum BSGP5+/6+ (BS) primer set on the basis of mismatch reduction to 48 genital HPV types. In addition, the method incorporates β-globin primers in the BS PCR as well as the standard GP PCR for internal DNA quality control. The sensitivity and performance of Luminex/BSGP5+/6+ was compared to that of standard RLB/GP5+/6+ with DNA extracted from cervical scrapings of a follow up population of 160 women with abnormal cervical cytology in Bogota, Colombia.
MATERIALS AND METHODOLOGY
Selection of Patients
We included in the study 160 adult women who were enrolled in a cervical cancer screening program and attending gynecologic services at different hospitals in the city of Bogota. Informed consent was signed by all participants, who voluntarily agreed taking part in the study for a period of eight months. Inclusion criteria were: women with history of borderline, mild or moderate dysplastic cytology (the criteria used for the cytologic diagnoses were those of The Bethesda System 2001) who were then referred to histopathology analysis, with the outcome of ‘normal’ or ‘abnormal’, in which case the lesions were classified according to the CIN grading system by the pathologist. Exclusion criteria were: pregnancy, previous history of biopsy-confirmed CIN (any grade) or HPV infection, as well as having received antineoplastic therapy, immunotherapy or HPV vaccination. All participants included in the study underwent subsequent exploratory colposcopy with collection of cervical scraping smears and biopsy samples at the site where the abnormal lesion was localized for HPV genotyping and histopathology analysis, respectively. According to the outcome of such analysis, the patients were diagnosed following the CIN grading system (CIN grades I, II and III), CC or as “negative pathology”. The histopathology findings were reviewed by a second pathologist, who was not blinded to the results. Those cases in which the diagnosis made by the two pathologists did not match were excluded from the study.
Collection and Processing of Cervical Samples
The cervical scrapings were collected at at the lesion site by colposcopy using cyto-brushes, which were immediately placed in sterile tubes containing 1 ml of nucleic acid-free phosphate buffer saline (PBS) supplemented with 0.05% thimerosal. All samples were subsequently treated with proteinase K (Bioline®) to a final concentration of 20 ug/ml, and incubated overnight at 37°C. Then, the proteinase K was inactivated at 95°C for 5 minutes, and the samples were centrifuged at 10.000 x g for 5 minutes and stored at -20°C until processed for DNA isolation. Cervical biopsies, taken at the site where the lesion was identified, were fixed with formalin and paraffin-embedded for histopathology analysis.
DNA Isolation from Cervical Scrapings
DNA was isolated from the samples treated with proteinase K in the previous section by a two-step phenol extraction and one-step chloroform-isoamyl alcohol extraction followed by ethanol precipitation. DNA pellets were resuspended in 0.2 ml of 10 mM Tris (pH 8.5) buffer and stored at -20°C until further analysis.
Quality Assessment of DNA
Quality of DNA was confirmed by electrophoresis and by PCR with primers PC03/PC04, which amplify a 110 bp fragment of the ß-globin gene [17Arias Y, Carrillo E, Aristizábal F. Plasma DNA restoration for PCR applications J Clin Pathol 2007; 60: 952-4.]. The reaction mixture contained 1X PCR buffer, 3.5 mM MgCI2, dNTP mix with 200 µM of each, PC03 and PC04 at 1 µM each, 1.25 U of Taq polymerase (Promega®) and distilled water to a total reaction volume of 25 µL. The reactions were run on the My CyclerTM Thermal Cycler (BioRad) using the following conditions: 4 minutes at 94 º C, followed by 40 cycles of amplification 94°C for 1 minute, 55ºC for 1 minute and 72ºC for 1 minute, and extension end at 72°C for 4 minutes. Lymphocyte DNA was used as positive control, and distilled water as negative control. The PCR products were visualized by gel electrophoresis on 1% agarose gels stained with ethidium bromide to identify the amplified 110pb band. The electrophoresis was run for one hour at 100 volts in TBE buffer using a BioRad Power PAC 3000 and visualized with a gel scanner with Quantity One software from BioRad.
HPV Genotyping by RLB
For all samples positive for the ß-globin amplification, GP PCR was performed using the primers GP5+/5'-biotinylated GP6+ as described elsewhere [15Van Den Brule A, Pol R, Fransen-Daalmeijer N, Schouls L, Meijer C, Snijders P. GP5+/GP6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes J Clin Microbiol 2002; 40: 779-87.]. The reaction mixture contained 10 µL of sample, 1X PCR buffer, 3.5 mM MgCI2, dNTPs mix (200 µM of each deoxynucleoside triphosphate), GP5+ and 5'-biotinylated GP6+ primers at 1 µM each, 1.25 U of Taq polymerase (Promega®) and sterile distilled water to a total reaction volume of 50 µL. Amplification was carried out in a thermocycler MyCyclerTM (Biorad) and PCR conditions were: one step of denaturation at 94°C for 4 minutes, followed by 40 amplification cycles each with the following steps: denaturation at 94°C for 1 minute, annealing at 40°C for 2 minutes and elongation at 72°C for 1½ minutes. A final elongation step was prolonged by 4 minutes. As positive control we used DNA extracted from the cervical cancer cell lines HeLa and SiHa kindly provided by the laboratory of Pharmacogenetics from the National University of Colombia.
Detection was performed using an Immunetics Minibloter® 45. This system uses membranes with carboxyl groups that are activated with 16% (w/v) EDAC (1-ethyl-3-(-3-dimethylamininopropyl) carbodiimide) to covalently bind oligonucleotides coupled to amino groups, which detect 37 types of HPV types as reported previously by Van den Brule [15Van Den Brule A, Pol R, Fransen-Daalmeijer N, Schouls L, Meijer C, Snijders P. GP5+/GP6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes J Clin Microbiol 2002; 40: 779-87.]. Subsequently, the membrane is rotated 90 degrees, for hybridization with denatured PCR products at 42ºC for 45 minutes, followed by two washes at 52 º C for 10 minutes with 2X SSPE (1X SSPE contains 0.18 M NaCl, 10 mM NaH2PO4 and 1 mM EDTA pH 7.4, Invitrogen) and 0.5% SDS. Then, the membrane was incubated with streptavidin-peroxidase conjugate (Roche®) (diluted 1:4000) for 45 minutes at 42°C, followed by two washes in 2X SSPE and 0.5% SDS for 10 minutes at 42 º C and two washes with 2X SSPE for 5 minutes at room temperature. Detection of specific hybridization was performed using ECL (GE Healthcare UK, Amersham) followed by exposure overnight of autoradiography film (Amersham Hyperfilm ™ ECL). The film was developed using a Medical Film Processor Model SRX-101 Konica Minolta Medical & Graphic, Inc.
Multiplex HPV Genotyping (MPG) and Estimation of Viral Load
Multiplex BSGP5+/GP6+ PCR-MPG was carried out with primers reported previously by Schmitt et al., 2008 [11Schmitt M, Dondog B, Waterboer T, Pawlita M. Homogeneous amplification of genital human alpha papillomaviruses by PCR using novel broad-spectrum GP5+ and GP6+ primers J Clin Microbiol 2008; 46: 1050-9.]. The assay used nine sense primers BSGP5+ at 200 nM and 3 antisense primers 5'-biotinylated-BSGP6+ at 400 nM to amplify a ~150 bp fragment of the viral L1 ORF, as well as two additional primers (MS3/MS10) at 300 nM to amplify the ß-globin gene, which was used as DNA integrity control [18Dunbar SA, Vander Zee CA, Oliver KG, Karem KL, Jacobson JW. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the luminex LabMAP system J Microbiol Methods 2003; 53: 245-52.]. The multiplex Luminex hybridization assay enabled the detection of 27 mucosal HPV-genotypes: i) high risk (HR)-HPV types: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68; ii) possibly HR-HPV types: 26, 53, 67, 70, 73, 82; low risk (LR)-HPV types: 6, 11, 30, 43, 44 and 69. In addition, the β-globin product was amplified in each sample.
Oligonucleotide probes with an amino group at the 5' end for each of the 27 HPV types were coupled to carboxylated beads using the carbodiimide procedure described by Schmitt et al., 2006 [16Schmitt M, Bravo I, Snijders P, Gissmann L, Pawlita M, Waterboer T. Bead-Based Multiplex Genotyping of Human Papillomaviruses J Clin Microbiol 2006; 44: 504-12.]. For each combination of probes and beads we used 2.5 million carboxylated beads in 25 µL of 0.1 M2-(N-morpholino) ethanesulfonic acid (MES), pH 4.5, with 200 µg of N-(3-dimethylaminopropyl)-N-ethyl-carbodiimide (EDC) and 400 pmol of probe, the mixture was incubated in the dark under stirring for 30 minutes, the addition of EDC and incubation was repeated once, then the beads were washed with 0.5 ml of 0.2 g/L of Tween 20 and once with 0.5 ml of 1.0 g/L SDS. Finally, they were stored at 4 °C in TE solution. Hybridization was performed in 96 well plates with 10 µL of PCR product in 33 µL of hybridization solution (0.15 M of tetramethylammonium chloride (TMAC), 75 mM Tris-HCl, 6 mM EDTA, 1.5 g/L Sarkosyl, pH 8.0) to which 2000 bead-coupled probes were added. This mixture was incubated at 95°C for 10 minutes and immediately placed on ice for 2 minutes, then it was placed at 41°C for 30 minutes under stirring, then the samples were transferred to a wash plate with filter bottom (Millipore, Bedford, MA) pre-equilibrated with blocking solution (PBS with 0.2 g/L Tween-20 and 2.0 M TMAC), and washed with blocking solution to eliminate non-hybridized DNA in a vacuum wash station (Millipore). Subsequently, biotinylated PCR products were stained with streptavidin-R-phycoerythrin conjugate (Molecular Probes) diluted 1:1600 in 2.0 M TMAC, 75 mM Tris-HCl, 6 mM EDTA, 1.5 g/L Sarkosyl, 1.0 g/L casein, pH 8.0 and incubated 20 minutes at room temperature with shaking. After washing three times with blocking solution, the beads were analyzed in a Luminex 100 reader (Luminex Corp.), which uses two lasers, one that recognizes the bead set by the internal bead color and another to quantify the reporter fluorescence on the bead. The results were expressed in median fluorescence intensities (MIF) of ≥100 beads per sample [11Schmitt M, Dondog B, Waterboer T, Pawlita M. Homogeneous amplification of genital human alpha papillomaviruses by PCR using novel broad-spectrum GP5+ and GP6+ primers J Clin Microbiol 2008; 46: 1050-9., 16Schmitt M, Bravo I, Snijders P, Gissmann L, Pawlita M, Waterboer T. Bead-Based Multiplex Genotyping of Human Papillomaviruses J Clin Microbiol 2006; 44: 504-12., 18Dunbar SA, Vander Zee CA, Oliver KG, Karem KL, Jacobson JW. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the luminex LabMAP system J Microbiol Methods 2003; 53: 245-52., 19Louie KS, de Sanjose S, Diaz M, et al. Early age at first sexual intercourse and early pregnancy are risk factors for cervical cancer in developing countries Br J Cancer 2009; 100: 1191-7.].
As control, plasmid standards from the WHO panel II [20Eklund C, Zhou T, Dillner J. for the WHO Human Papillomavirus Laboratory Network. Global proficiency study of human papillomavirus genotyping J Clin Microbiol 2010; 48: 4147-55.] with 100 and 1,000 copies for 16 distinct HPV types per 100ng human placental DNA were amplified and served to estimate the HPV viral load. MFI values obtained with 1,000 HPV genome copies of the respective HPV types per 100 ng of human placental DNA served as cutoff for defining low (<cutoff) and high viral load (>cutoff) groups.
Statistical analysis
The demographic data was analyzed using Statistix 9 software. The genotyping results were analyzed using Excel®. For each probe, MFI values in reactions with no PCR product added to the hybridization mixture were considered background values. Reactions with net MFI values above 5 were defined as positive reactions. Net MFI values were computed by subtraction of 1.1 times the median background value. For all probes, this cutoff value was above the mean background plus three times the standard deviation.. P values of <0.05 were considered statistically significant.
RESULTS
Population Under Study
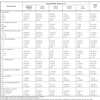
Inclusion and exclusion criteria for selection of the study population are defined in the Methods section. All participants had a history of abnormal cervical cytology (atypical squamous cells, any grade, according to The Bethesda System 2001) and underwent subsequent colposcopy and histopathology analysis, which led to their classification into one of the five groups: negative pathology, CIN I, CIN II, CIN III and CC. The Table 1 resumes the socio-demographic and anamnestic characteristics of the study population distributed by the grade of the lesions found in the histopathologic analysis. Nearly 51% of the participants in the study were under the age of 34 years, with ~52% belonging to socioeconomic levels 1 or 2, classified according to the criteria of the Secretary for District Planning and the National Administration Department for Statistics (DANE, Decreto 304, 2008). Nearly half of the population (50.6%) had their first sexual intercourse under the age of 18, and about 70% had 1-2 sexual partners. The majority of women participating in the study had children (85.6%), and their first delivery was in most cases (87.6%) previous to the age of 24. A large number (56.9%) never used contraceptive methods. About 63% had no family history of cancer. A family history of CC was infrequent in all types of CIN lesions (Table 1) and even absent in the CC patients. However, a family history of other types of cancer was found in a significant number of women in all groups with a maximum in CIN II (43.8%).
Histopathologic Characteristics
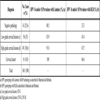
The distribution of cervical cancer and its precursor lesions among the study population is summarized in Table 2. Of the 160 study participants 41 (25.6%) did not show any detectable cervical lesions in the histopathology analysis. The remainder 119 cases were diagnosed of cervical lesions at different stages: 56 (35%) with low-grade cervical lesions (CIN I), 49 (30.6%) with high-grade cervical lesions (CIN II and CIN III) and 14 women (8.8%) were diagnosed of CC (either in situ or invasive carcinoma).
In a previous study, Wright and colleagues estimated the sensitivity of the cytology screening to be about 60.7% [21Wright TC, Denny L, Kuhn L, Pollack A, Lorincz A. HPV DNA testing of self-collected vaginal samples compared with cytologic screening to detect cervical cancer JAMA 2000; 283: 81-6.], with a trend to increase with higher degree of dysplasia. In our study, the sensitivity of the Pap smear was determined to be 69,6% for CIN I, 40% for CIN II, 63% for CIN III and 78,6% for CC.
HPV Prevalence
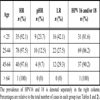
The majority of CIN and CC cases are linked to infection by the so-called high-risk HPVs, a subset of oncogenic HPV types with mucosal tropism [6Kisseljov FL, Kisseljova NP, Kobzeva VK, et al. State of human papilloma virus DNA in cervical carcinomas Mol Biol 2001; 35: 399-404., 7Cid-Arregui A. Therapeutic vaccines against human papillomavirus and cervical cancer Open Virol J 2009; 3: 67-83.]. We used both multiplex PCR and Luminex xMAP technology as well as reverse line blot (RLB) analysis for detection and genotyping of the HPV types 16 and 18, the most prevalent in CC, in DNA isolated from cervical smears of all the patients participating in the study. As expected, these two viral types were found at high frequencies in all five histopathologic groups. With the Luminex xMAP technique HPV16 and HPV18 were detected in separate or combined infections in 80.5% of women with negative histopathology diagnosis, 83.9% of low-grade lesions (CIN I), 91.8% of high-grade lesions (CIN II and CIN III) and 100% of CC (Table 2, middle column). In contrast, the RLB analysis detected the presence of these virus types at markedly lower frequencies in all groups (Table 2, right column), suggesting that, in our hands, this technique is less sensitive than the Luminex xMAP method. Furthermore, in those samples positive for HPV by both methods there was a 67.2% coincidence in the HPV type(s) detected (p<0.05). Therefore, Luminex xMAP was chosen to detect up to 27 mucosotropic HPV types in cervical scrapings of all the women included in the study. Table 3 summarizes the prevalence of the HPV types that were detected in the five histopathologic groups of our study population.
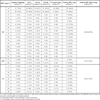
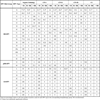
Using Luminex xMAP one or more HPV types were demonstrated in the cervical smears of all 160 participants in the study. Table 3 shows the 27 HPV types that were detected. Of these, 16 were high-risk (HR), 2 possibly high-risk (pHR) and 9 low-risk types commonly found in genital warts and low-grade cervical lesions [22Muñoz N, Bosch FX, de San Jose S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer N Engl J Med 2003; 348: 518-27.]. The data in Table 3 reflects the HPV types detected either as unique or double/multiple infections. HPV16 was by far the most prevalent virus (overall prevalence 84.4%), ranging from 75.6% in the negative pathology group to 100 in CC. The second most common virus type was HPV18 (overall prevalence 84.4%), with 53.6% in the negative pathology group and over 60% in all CIN groups, the lowest prevalence being in the CC group (28.5%). Among the remainder HR-HPV types, only five were found in CC: HPV45, 52, 56, 68 and 69, although all HR-HPV types detected in our study were also present in the negative pathology and/or the CIN groups. Of the two pHR-HPV types detected (HPV53 and 66) only HPV53 was found in CC, albeit HPV66 was present in all other groups with a relatively high prevalence (about 10-15%). Among the LR-HPV types only four types were found in CC (HPV6, 42, 69 and 70) of which HPV6 showed the highest prevalence (21.4%).
The accuracy of the Luminex genotyping was further confirmed by analyzing the expression of early genes (E6 and E7) of the most prevalent HR-HPVs detected with this technique (HPV types 16, 18, 31, 33, 45, 51, 52, 56 and 58). To this end, we isolated total RNA from biopsy material of the patients positive for these virus types and, after reverse transcription and DNase I treatment, analyzed them by quantitative real time PCR (QRT-PCR) using specific primers for E7 or E6/E7 transcripts. The results validated those obtained with Luminex, since all samples identified as positive for the specific virus types by Luminex amplified their respective transcripts by QRT-PCR (data not shown).
Single Versus Multiple Infections
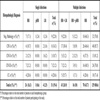
The Table 4 shows the frequencies of infection with HR, pHR and LR-HPVs found in the five histopathologic groups as single or multiple events. Overall, the frequency of multiple infections (129/160 cases, 80.6%) was about 4-fold higher than that of single infections (31/160, 19.4%), a trend that was conserved in the five groups of patients. HR-HPV types were the most prevalent in all cases of infection with a unique virus, the highest prevalence being in the CC group (28.6%). In contrast, single infections with LR-HPVs were rare and were not observed at stages above CIN II. There was just one case of single infection by pHR-HPV in the negative pathology group. In the samples infected with more than one HPV type, the combination of two or more HR-HPVs was the most frequent (70/160, 43.7%), followed by HR+LR infections (43/160, 26.9%). The combination of HR+pHR was rare (16/160, 10%) (Table 4). Within the CC group, HR+LR-HPV infections were more frequent (5/14, 35.7%) than infections with multiple HR-HPV types (4/14, 28.6%). However, such differences were not statistically significant (p>0.1, one way ANOVA).
HPV Prevalence by Age
Analysis of the prevalence of HR-HPVs by age showed an overwhelming presence of these viruses in all age groups, with over 97% of women above the age of 25 being positive (Table 5). Among the HR-HPVs, HPV16 and 18 were found in 80-90% of cases, as single or multiple infections. The LR-HPVs showed much lower frequency rates, with a maximum in the group less than 25 years old (42.1%). Likewise, pHR-HPVs had their maximal prevalence in the same age group, but with a lower frequency rate (23.7%).
Viral Load
The viral load was estimated in the DNA isolated from the cervical scrapings by calculating the HPV copy number against standards. As can be appreciated in the Table 6, the samples infected with HR-HPVs, especially HPV types 16, 18, but also types 31, 33, 52 and 58, had an overall tendency towards a stronger viral load, which correlated with the degree of dysplasia. However the differences seen were not statistically significant. For the two most prevalent HR-HPV types, 16 and 18, the highest copy numbers were found in the group diagnosed of CIN II in which about 80% had more than 50 viral copies. Interestingly, the viral load of these two viruses decreased in CIN III and CC (p<0.05). In contrast, most samples infected with LR-HPVs contained less than 50 viral copies, with the exception of those infected with HPV6, a significant number of which carried higher numbers of copies, particularly in CIN I and II lesions (p<0.05). Of the two pHR-HPVs that were detected, HPV53 was in the range below 50 copies, while HPV66 showed a tendency to higher viral load, largely in the CIN I group, but the difference was not statistically significant.
DISCUSSION
In this multicenter study we analyzed the prevalence of a subset of genital HPVs in 160 patients with abnormal cervical cytology, who were subsequently examined and diagnosed by the pathologists and classified into five groups ranging from absence of appreciable pathology to cervical cancer. These women agreed to participate in our study and donated a part of the cervical scrapings taken for diagnostic purposes to be used for HPV detection and typification. It has been estimated that 99.7% of invasive CC cases worldwide are associated with infection by HPV [8Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide J Pathol 1999; 189: 12-9.]. The HPV types with genital tropism have been divided into three groups: HR, pHR and LR. Infection with HR-HPV types is an essential risk factor for progression of cervical dysplastic lesions to CC. Hence, HPV detection and typification has become a routine tool in cervical screening, epidemiological studies, as well as for monitoring CIN and CC progression. HPV detection provides diagnostic and prognostic value, which should improve the relatively low sensitivity of cytology, estimated to be about 60.7% [Ref. 21 and our own results].
A first aim of our study was to compare the sensitivity of two methods of HPV detection: RLB, a quick and easy method designed to identify HPV genotypes [15Van Den Brule A, Pol R, Fransen-Daalmeijer N, Schouls L, Meijer C, Snijders P. GP5+/GP6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes J Clin Microbiol 2002; 40: 779-87.], and Luminex xMAP, a highly sensitive automated technique developed recently [11Schmitt M, Dondog B, Waterboer T, Pawlita M. Homogeneous amplification of genital human alpha papillomaviruses by PCR using novel broad-spectrum GP5+ and GP6+ primers J Clin Microbiol 2008; 46: 1050-9.], which can detect 27 HPV types simultaneously. Our results show that the sensitivity of Luminex xMAP for HPV detection is remarkably higher than that of RLB (see Table 2). This was shown for HPV16 and 18, which are the most prevalent viruses found in CC, but was also confirmed for other HPV types with lower prevalence (data not shown). Overall, the frequency of detection with Luminex xMAP of these two viral types in the negative pathology and low-grade groups was significantly higher than with RLB (2.4-fold and 1.9-fold, respectively), and was also higher in the high-grade and CC groups (about 1.4-fold). The lower sensitivity of RLB cannot be explained by differences in the quality of the samples, which were the same for both RLB and Luminex, but may reflect the higher sensitivity of the probes used and the assay as has been reported previously (16).
The application of Luminex xMAP technology to the detection of HR, pHR and LR-HPV types in cervical scrapings of our study population showed a considerable prevalence of HR-HPV (96.25%), comparable to that reported previously [22Muñoz N, Bosch FX, de San Jose S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer N Engl J Med 2003; 348: 518-27., 23Murillo R, Molano M, Martínez G, Mejía JC, Gamboa O. HPV prevalence in Colombian women with cervical cancer: implications for vaccination in a developing country Infect Dis Obstet Gynecol 2009; 653598.] (Table 3). Overall, the most prevalent viral types were HPV16 (84.4%) and HPV18 (60.6%), followed by HPV56 (16.9%), HPV52 and 66 (both 10.6%). These values are higher than those reported previously by Clifford et al. in cytologically normal women [24Clifford GM, Gallus S, Herrero R, et al. Worldwide distribution of human papillomavirus types in cytologically normal women in the International Agency for Research on Cancer HPV prevalence surveys: a pooled analysis Lancet 2005; 366: 991-8.]. Such discrepancy may be explained by the fact that our study population was selected among women with a history of abnormal cytology and thus more likely to harbor HPV infections. Surprisingly, the high prevalence of HPV18 in CIN I, II and III, which was over 60%, dropped to 28.5% in CC. Other HR-HPVs (types 31, 33, 35, 39, 51, 68a, 68b, 72 and 88), which had a significant prevalence in CIN lesions, were absent in CC. The same was observed for the pHR-HPV type 66. This phenomenon suggests that these HPV types may act as companions/helpers of more prevalent HPVs during the precancerous CIN stages, but cannot overcome the oncogenic capability of the later, in particular HPV16, are less prone to integrate in the host genome and disappear. It is noteworthy that also LR-HPVs were detected in advanced CIN lesions, though only four types (HPV 6, 42, 69 and 70) were found in CC, of which HPV6 was the most prevalent (~20%).
A majority of the participants in the study (~80%) were positive for several HPV types (Table 4), indicating that multiple infections are a common phenomenon. Indeed, multiple infections have been detected with the Luminex technique in up to 58% of DNA samples from cervical scrapings of women participating in a population-based cervical screening trial in Mongolia [25Schmitt M, Dondog B, Waterboer T, Pawlita M, Tommasino M, Gheit T. Abundance of multiple high-risk human papillomavirus (HPV) infections found in cervical cells analyzed by use of an ultrasensitive HPV genotyping assay J Clin Microbiol 2010; 48: 143-9.]. A recent study in a Colombian population found 62.5% of multiple infections among HPV-infected women by conventional PCR [26Soto-De Leon S, Camargo M, Sanchez R, et al. Distribution patterns of infection with multiple types of human papillomaviruses and their association with risk factors PLoS One 2011; 6: e14705.]. The higher frequency of multiple infections found in our study as compared to this may reflect the higher sensitivity of the Luminex assay.
Multiple infections with only HR-HPV types were the most frequent, followed by the combination of HR and LR-HPV and of HR with pHR-HPV types. Interestingly, in the CC group HR+LR-HPV infections were slightly more frequent (35.7%) than infections with HR-HPV types (28.6%), the most prevalent being HPV6. This result suggests that the LR-HPVs persist in cancer lesions possibly as commensals, since all CIN III were negative for HPV6. A larger cohort of patients should be analyzed to confirm such high frequency of non-oncogenic LR-HPVs in CC. As could be expected, in those CIN III and CC cases in which a unique HPV type was detected this was a HR-HPV. Our results are in contrast with those of Muñoz and colleagues, who reported frequencies of 91.9% and 8.1% for single and multiple infections, respectively [22Muñoz N, Bosch FX, de San Jose S, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer N Engl J Med 2003; 348: 518-27.]. In that study, HPV detection and typing was made by a PCR assay using MY09/MY11 and GP5+/6+ primers. Therefore, the discrepancy can be explained by the different sensitivity of the techniques used in both studies.
The distribution of infections with the different HPV types by age showed a remarkably high prevalence of HR-HPVs in all age groups. HPV types 16 and 18 were the most prevalent, even in younger women. These results are in support of vaccination against those viruses as early as possible.
The viral load data showed a trend of HR-HPVs towards higher copy numbers correlating with the degree of dysplasia. For lesions infected with HPV16 and 18, of which a larger number of cases was analyzed, CIN II appeared to be the lesion with higher viral load. A possible explanation for this may be the episomal state of the virus at that stage, which may allow for accumulation of higher numbers of copies, what may not happen after the virus integrates into the host genome at later stages. Viral load of LR-HPVs was in the lower range, except for HPV6, which showed a tendency to higher copy numbers in CIN lesions.
Altogether, these results indicate that HPV16 may play a more essential role in the development of CC in Colombian women than previously thought. The frequency of HPV16 was previously reported to be nearly 50% in HPV-positive women in Colombia [26Soto-De Leon S, Camargo M, Sanchez R, et al. Distribution patterns of infection with multiple types of human papillomaviruses and their association with risk factors PLoS One 2011; 6: e14705.]. In contrast, in our study population we found a significantly higher frequency of HPV16 (84.4%). Again, the higher sensitivity of our Luminex assay compared to the standard PCR used in that study may account for the difference, although geographical and socioeconomic factors related with the two populations might have also contributed to the higher HPV16 frequency observed in ours.
Furthermore, since HPV16, the most oncogenic HPV known, was detected in all CC cases, the presence of other HPV types in the same lesions may represent concomitant infections. It remains to be determined to which extent each virus contributes to the oncogenic transformation. Further, it needs to be clarified whether these co-infections occur in the same or in different cervical epithelial cells needs to be clarified, for instance by in situ hybridization analysis of biopsy samples using type-specific probes.
ACKNOWLEDGEMENTS
We are grateful to the departments of Gynecology-Obstetrics and Pathology of Dinamica-IPS, Hospital San Ignacio, Fundación Santa Fe, Hospital Engativá and clinical teleton. We are also thankful to the Faculty of Medicine of La Sabana University for financial support.