- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
A Cell-Based Protein-Protein Interaction Method Using a Permuted Luciferase Reporter
Haifeng Eishingdrelo*, 1, Jidong Cai 2, Paul Weissensee 2, Praveen Sharma 2, Michael J Tocci 2, Paul S Wright 3
Abstract
We have developed a novel cell-based protein-protein interaction assay method. The method relies on conversion of an inactive permuted luciferase containing a Tobacco Etch Virus protease (TEV) cleavage sequence fused onto protein (A) to an active luciferase upon interaction and cleavage by another protein (B) fused with the TEV protease. We demonstrate assay applicability for ligand-induced protein-protein interactions including G-protein coupled receptors, receptor tyrosine kinases and nuclear hormone receptors.
Article Information
Identifiers and Pagination:
Year: 2011Volume: 5
First Page: 122
Last Page: 128
Publisher Id: CCGTM-5-122
DOI: 10.2174/1875397301105010122
Article History:
Received Date: 19/5/2011Revision Received Date: 17/8/2011
Acceptance Date: 18/8/2011
Electronic publication date: 30/11/2011
Collection year: 2011
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the BioInvenu Corporation, USA, East Hanover, NJ, USA; Tel: 973-585-6777; Fax: 973-585-6776; E-mail: haifeng.eishingdrelo@bioinvenu.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 19-5-2011 |
Original Manuscript | A Cell-Based Protein-Protein Interaction Method Using a Permuted Luciferase Reporter | |
INTRODUCTION
A variety of mammalian cell based protein-protein interaction methods have been developed. The methods include imaging interaction of fluorescence proteins [1Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis Nat Biotechnol 2003; 21: 539-45.], fluorescence or bioluminescence resonance energy transfer [2Ramsay D, Kellet E, McVey M, Rees S, Milligan G. Homo- and hetero-ologomeric interactions between G-protein-coupled receptors in living cells monitored by two variants of bioluminescence resonance energy transfer (BRET): hetero-oligomers between receptor subtypes form more efficiently than between less closely related sequences Biochem J 2002; 365: 429-0.,3Ai HW, Hazelwood KL, Davidson MW, Campbell RE. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors Nat Methods 2008; 5: 401-3.], protein-fragment or enzyme-fragment complementation [4Wehr MC, Laage R, Bolz U, et al. Monitoring regulated protein-protein interactions using split TEV Nat Methods 2006; 3: 985-3.-7Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia luciferase Nat Methods 2006; 3: 977-.], and interaction-induced transcriptional activated reporters [8Barnea G, Strapps W, Herrada G, et al. The genetic design of sig-naling cascades to record receptor activation Proc Natl Acad Sci USA 2008; 105: 64-9.]. Here we describe a novel method based on an inactive permuted protein which can be activated by a protease to generate stable and sensitive signals for monitoring protein-protein interactions in mammalian cells. The basic protein-protein interaction assay design consists of two components: an inactive permuted luciferase containing a Tobacco Etch Virus (TEV) protease cleavage sequence fused to protein A, and protein B fused to the protease TEV [9Kapust RB, Tozer J, Copeland TD, Waugh DS. The P1’ specificity of tobacco etch virus protease BBRC 2002; 294: 949-55.]. Upon interaction between protein A and B, inactive permuted luciferase is cleaved and active luciferase is reconstituted. The luciferase signals should be specific and sensitive for specified protein A and B interaction. We named this cell-based protein-protein interaction method as LinkLight assay technology, meaning light is generated upon protein A interacting with protein B (Fig. 1a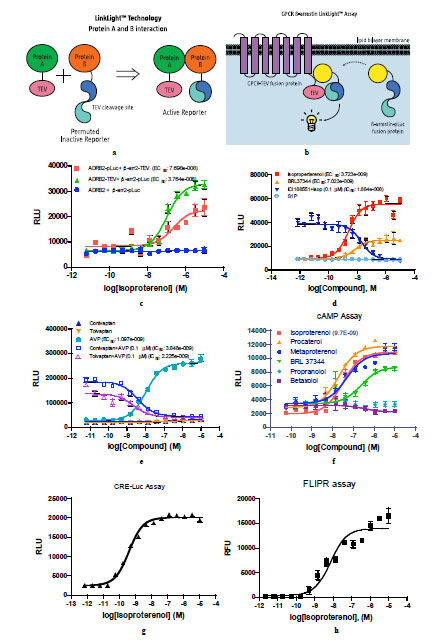 ). The LinkLight assay does not involve transcription and translation, does not need long exposure times with compounds, and therefore, has reduced off-target signals. The LinkLight assay is different from bioluminescence resonance energy transfer (BRET) and fluorescence resonance energy transfer (FRET) assays which depend on the degree of the spectral overlap, the relative orientation, and the distance between the donor and acceptor. The method is also different from protein-fragment complementation assays (PCA) or enzyme-fragment complementation assays (ECA) in which fragment spontaneous self-complementation could produce high background signals, especially when the proteins are over-expressed and the two fragments have high complementation affinity. Based on the assay principal, the method described here should generate stable signals for both transient and stable protein-protein interactions.
). The LinkLight assay does not involve transcription and translation, does not need long exposure times with compounds, and therefore, has reduced off-target signals. The LinkLight assay is different from bioluminescence resonance energy transfer (BRET) and fluorescence resonance energy transfer (FRET) assays which depend on the degree of the spectral overlap, the relative orientation, and the distance between the donor and acceptor. The method is also different from protein-fragment complementation assays (PCA) or enzyme-fragment complementation assays (ECA) in which fragment spontaneous self-complementation could produce high background signals, especially when the proteins are over-expressed and the two fragments have high complementation affinity. Based on the assay principal, the method described here should generate stable signals for both transient and stable protein-protein interactions.
MATERIALS AND METHODOLOGIES
Plasmid Construction
Human G-protein coupled receptor (GPCR) DNA fragments without stop codon were generated by polymerase chain reaction (PCR) for subsequent subcloning. A flexible peptide sequence (G4S)3 was used to link GPCR and TEV protease. The tobacco TEV fragment contain 240 amino acid sequence was obtained by PCR using tobacco etch virus DNA clone (ATCC # 45035) as the template. PCR primers: CTCGAGGGAGAAAGCTTGTTTAAGGGAC (5’ primer) and GGGCCCCTATTGCGAGTACACCAATTCATTC (3’ primer) contain introduced Xho I and Apa I restriction enzyme sites to facilitate cloning. GPCR-TEV fusion genes were then subcloned into the pcDNA3.1-zero or –hyg vector. Firefly luciferase DNA fragments corresponding to amino acid sequence 2 to 233 and 234 to 550 were generated by PCR using pCRE-Luc (GeneBank accession: AF053461.1) as the template and high-fidelity DNA polymerase (Invitrogen). An inactive permuted luciferase (pLuc) was constructed by using a TEV protease cleavage sequence encoding ENLYFQR was used as a linker to link the C luciferase fragment containing amino acids 234 to 550 in front of N fragment containing amino acids 2 to 233. The permuted luciferase was then fused to the C-terminus of β-arrestin 2 and subcloned into the pcDNA 3.1 vector (Invitrogen). Similarly, nuclear hormone receptor (NHR)-TEV and receptor tyrosine kinase (RTK)-TEV constructs were generated by replacing GPCR DNA fragment in the GPCR-TEV construct with either NHR or RTK DNA fragments by standard molecular cloning method. NCOA1-pLuc was constructed by replacing β-arrestin 2 DNA fragment in β-arrestin 2-pLuc construct with DNAs encoding NCOA1 nuclear localization signal sequence and NCOA1 amino acid sequence from 630 to 760. PTB-pLuc was constructed by fusing DNA fragment encoding PTB domain of Src homology 2 domain containing transforming protein 1 (SHC1) in front of permuted luciferase sequence. All constructs were verified by DNA sequencing.
Stable Cell Line Generation
CHO, HEK293, and U-2 OS cells were routinely maintained and passaged in standard recommended medium with Pen/Strep (Invitrogen). Monoclonal cell lines stably expressing β-arrestin-permuted luciferase fusion protein (HEK__β-arr2-pLuc and U2OS__β-arr2-pLuc) were selected with G418 250 ìg/ml for HEK293 cells or Hygromycin B 500 µg/ml for U2OS cells. Monoclonal U2OS_PTB-pLuc and U2OS_NCOA1-pLuc were selected with Hygromycin B 500 µg/ml.
Chemicals
All the chemicals were purchased from Sigma-Aldrich (USA) and Tocris Biosciences.
LinkLight Assay
Cells stably expressing or transiently expressing GPCR-TEV and β-arr2-pLuc genes were plated into a 384-well opaque assay plate (Becton Dickinson) at 10,000 cells per well. Cells were incubated overnight, and the culture medium was replaced with serum-free medium and compounds were added into the wells. Compounds were added typically as dilutions into the culture medium from 10 mM DMSO stocks. After 90 min incubation equal volume of Bright-Glo luciferase detection reagent (Promega) or BrightLite (Perkin Elmer) detection reagent was then added to the wells and relative luminescence units (RLU) were recorded on either an EnVision or a TopCount plate reader (Perkin Elmer). Alternatively, after 90 min compound incubation, the culture medium was removed, and 15 μl of detection reagent was added to each well, then relative luminescent signals were recorded.
Cyclic AMP and CRE-Luc Assays
PathHunterTM cAMP kit (DiscoverX) was used in the cAMP assay. The assay was performed according to the manufacture’s instruction. For CRE-Luc assay, CHO cells stably expressing CRE-luciferase reporter gene were transfected with ADRB2-TEV construct and plated in 384-well plates. After 24 hour culture, the cells were treated with different concentrations of isoproterenol for 6 hours, and equal volume of Bright-Glo was added to each well and RLUs were recorded on an EnVision plate reader.
Flouorometric Imaging Plate Reader (FLIPR) Assay
HEK293 cells containing G protein Gα16 were seeded in 384-well plates with 10,000 cells per well and were transfected with GPCR-TEV constructs for 24 hours. Cells were then loaded with a calcium-sensitive fluorescent dye (FLIPR Calcium 3 assay kit, Molecular Devices). After 90 min incubation, compound was added to each well, and fluorescence signals were recorded on Fluorometric Imaging Plate Reader 384 (Molecular Devices).
RESULTS AND DISCUSSION
G-protein coupled receptor and β-arrestin interactions have been well documented [10Barak LS, Ferguson SS, Zhang J, Caron MG. A beta-arrestin/green fluorescent protein biosensor for detecting G protein-coupled receptor activation J Biol Chem 1997; 272: 27497-500., 11Shenoy SK, Lefkowitz RJ. Receptor-specific ubiquitination of beta-arrestin directs assembly and targeting of seven-transmembrane receptor signalosomes J Biol Chem 2005; 280: 15315-24.]. Biased ligands that selectively activate or inhibit G-protein vs. β-arrestin signaling pathways can have therapeutic benefits [12Violin JD, Lefkowitz RJ. B-Arrestin-biased ligands at seven-transmembrane receptors Trends Pharmacol Sci 2007; 28: 416-22.]. We used the GPCR-arrestin interaction as the model system for GPCR LinkLight assay development (Fig. 1b ). Inactive permuted luciferase (pLuc) was created by breaking the firefly luciferase coding region into two fragments, re-arranging the original fragment order, and re-connecting the fragments with a TEV protease cleavage site. The resulting permuted luciferase has no enzymatic activity. One particular inactive permuted luciferase was created by placing the firefly luciferase amino acid sequence 234 to 550 (C-fragment) in front of the amino acid sequence 2 to 233 (N-fragment). We chose the TEV cleavage sequence, ENLYFQR, for inclusion in the permuted luciferase gene construct. We evaluated two approaches for the assay development. First, we constructed ADRB2-TEV plasmid by fusing the TEV protease at the C-terminal of β-2 adrenergic receptor (ADRB2) and constructed β-arr2-pLuc plasmid by fusing the permuted luciferase (pLuc) at the C-terminal of beta-arrestin-2 (β-arr2). Second, we switched the fusion protein tags of the first design by fusing β-arrestin-2 with TEV and β-arrestin-2 with the pLuc. Transient expression of two sets of ADRB2 and β-arrestin-2 fusion gene constructs in HEK293 cells produced agonist isoproterenol-induced luciferase signals. Clearly, the first approach, in which ADRB2 tagged with TEV (ADRB2-TEV) and β-arrestin-2 tagged with the permuted luciferase (β-arr2-pLuc), exhibited the stronger signal and greater dynamic range (Fig. 1c
). Inactive permuted luciferase (pLuc) was created by breaking the firefly luciferase coding region into two fragments, re-arranging the original fragment order, and re-connecting the fragments with a TEV protease cleavage site. The resulting permuted luciferase has no enzymatic activity. One particular inactive permuted luciferase was created by placing the firefly luciferase amino acid sequence 234 to 550 (C-fragment) in front of the amino acid sequence 2 to 233 (N-fragment). We chose the TEV cleavage sequence, ENLYFQR, for inclusion in the permuted luciferase gene construct. We evaluated two approaches for the assay development. First, we constructed ADRB2-TEV plasmid by fusing the TEV protease at the C-terminal of β-2 adrenergic receptor (ADRB2) and constructed β-arr2-pLuc plasmid by fusing the permuted luciferase (pLuc) at the C-terminal of beta-arrestin-2 (β-arr2). Second, we switched the fusion protein tags of the first design by fusing β-arrestin-2 with TEV and β-arrestin-2 with the pLuc. Transient expression of two sets of ADRB2 and β-arrestin-2 fusion gene constructs in HEK293 cells produced agonist isoproterenol-induced luciferase signals. Clearly, the first approach, in which ADRB2 tagged with TEV (ADRB2-TEV) and β-arrestin-2 tagged with the permuted luciferase (β-arr2-pLuc), exhibited the stronger signal and greater dynamic range (Fig. 1c ). We speculate that the cytoplasmic environment, rather than proximity to the membrane is more favorable for the cleaved permuted luciferase fragments to reconstitute active luciferase. As a negative control, we co-transfected cells with β-arr2-pLuc and ADRB2 without TEV fusion protein constructs, no luciferase signals were observed. We then generated HEK293 cells stably expressing ADRB2-TEV and β-arr2-pLuc genes. The stable cells produced much larger signal dynamic ranges since we chose a stable β-arr2-pLuc reporter line with a low basal level signal while having high ligand-induced signal. Antagonist activity blocking agonist-induced signals can be easily detected. In addition, ligand sphingosine-1-phosphate for the endogenous S1P1 receptor in the HEK293 host cells did not produce detectable signals (Fig. 1d
). We speculate that the cytoplasmic environment, rather than proximity to the membrane is more favorable for the cleaved permuted luciferase fragments to reconstitute active luciferase. As a negative control, we co-transfected cells with β-arr2-pLuc and ADRB2 without TEV fusion protein constructs, no luciferase signals were observed. We then generated HEK293 cells stably expressing ADRB2-TEV and β-arr2-pLuc genes. The stable cells produced much larger signal dynamic ranges since we chose a stable β-arr2-pLuc reporter line with a low basal level signal while having high ligand-induced signal. Antagonist activity blocking agonist-induced signals can be easily detected. In addition, ligand sphingosine-1-phosphate for the endogenous S1P1 receptor in the HEK293 host cells did not produce detectable signals (Fig. 1d ), indicating that the endogenous receptor and its ligand did not interfere with the assay. Since class-A rhodopsin GPCRs can be divided into two subtypes based on β-arrestin-2 recruitment and receptor desensitization, we next evaluated the vasopressin V2 receptor (V2R) in the assay development. V2R, unlike ADRB2, which is a type A receptor that undergoes fast desensitization and dissociates from β-arrestin at the plasma membrane, is a type B receptor that slowly desensitizes and internalizes to endosomes with β-arrestins [13Oakley RH, Laporte SA, Holt JA, Barak LS, Caron MG. Association of beta-arrestin with G protein-coupled receptors during clathrin-mediated endocytosis dictates the profile of receptor resensitization J Biol Chem 1999; 274: 32248-57.]. HEK cells with co-stable expression of V2R-TEV and β-arr2-pLuc genes produced dose-dependent signals in response to agonist arg8-vasopressin (Fig. 1e
), indicating that the endogenous receptor and its ligand did not interfere with the assay. Since class-A rhodopsin GPCRs can be divided into two subtypes based on β-arrestin-2 recruitment and receptor desensitization, we next evaluated the vasopressin V2 receptor (V2R) in the assay development. V2R, unlike ADRB2, which is a type A receptor that undergoes fast desensitization and dissociates from β-arrestin at the plasma membrane, is a type B receptor that slowly desensitizes and internalizes to endosomes with β-arrestins [13Oakley RH, Laporte SA, Holt JA, Barak LS, Caron MG. Association of beta-arrestin with G protein-coupled receptors during clathrin-mediated endocytosis dictates the profile of receptor resensitization J Biol Chem 1999; 274: 32248-57.]. HEK cells with co-stable expression of V2R-TEV and β-arr2-pLuc genes produced dose-dependent signals in response to agonist arg8-vasopressin (Fig. 1e ), indicating that the GPCR beta-arrestin interaction assay can be applied to both type A and type B receptors.
), indicating that the GPCR beta-arrestin interaction assay can be applied to both type A and type B receptors.
To ascertain whether the protein interactions detected in GPCR LinkLight assay represented biologically functional interactions, we investigated G-protein signaling with the TEV tagged GPCRs. We performed cAMP assays in HEK293 cells stably expressing ADRB2-TEV and β-arr2-pLuc with an array of ligands. The results of the cAMP assay (Fig. 1f ) were comparable to the LinkLight assay results. ADRB2 is Gs coupled receptor and increases cAMP levels in response to agonist stimulation. CHO cells containing a CRE-Luc reporter gene were transiently transfected with the ADRB2-TEV fusion protein construct. We observed ligand-induced CRE-Luc reporter signals (Fig. 1g
) were comparable to the LinkLight assay results. ADRB2 is Gs coupled receptor and increases cAMP levels in response to agonist stimulation. CHO cells containing a CRE-Luc reporter gene were transiently transfected with the ADRB2-TEV fusion protein construct. We observed ligand-induced CRE-Luc reporter signals (Fig. 1g ). Again, TEV tagged GPCR did not alter the G-protein signals. In addition, when HEK293 cells containing a promiscuous Gα16 protein transiently expressed the ADRB2-TEV fusion protein, ligand-induced Ca2+ mobilization signals were observed in the FLIPR assay. The experiment yielded the expected EC50 values (Fig. 1h
). Again, TEV tagged GPCR did not alter the G-protein signals. In addition, when HEK293 cells containing a promiscuous Gα16 protein transiently expressed the ADRB2-TEV fusion protein, ligand-induced Ca2+ mobilization signals were observed in the FLIPR assay. The experiment yielded the expected EC50 values (Fig. 1h ). The results indicate that the GPCR-TEV fusion protein does not interfere with G-protein signaling. The EC50 values of the four different assay formats tested using the ADRB2-TEV construct are summarized in Table 1.
). The results indicate that the GPCR-TEV fusion protein does not interfere with G-protein signaling. The EC50 values of the four different assay formats tested using the ADRB2-TEV construct are summarized in Table 1.
To assess general applicability of the conceptual assay design for other protein-protein interactions, we investigated nuclear hormone receptor (NHR) LinkLight assays (Fig. 2a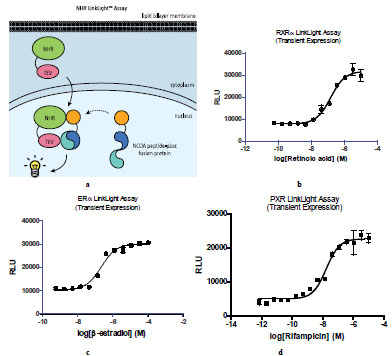 ). Once activated, nuclear hormone receptors translocate into the nucleus and interact with nuclear receptor co-activator-1 (NCOA1) to form the transcriptional complex [14Darimont BD, Wagner RL, Apriletti JW, et al. Strucutre and specificity of nuclear receptor-coactivator interactions Genes Dev 1998; 2: 3343-56.]. We tagged retinoid X receptor α (RXRα) with TEV at the C-terminal (RXRα-TEV) and fused a nuclear hormone interaction domain of NCOA1 with the permuted luciferase (NCOA1-pLuc). We expected that active luciferase signals would be generated once RXRα-TEV and NCOA1-pLuc fusion protein make contact in the nucleus. In order to test this, we generated U2OS cells stably expressing NCOA1-pLuc and designated these as U2OS_NCOA1-pLuc cells. When U2OS_NCOA1-pLuc cells were transiently expressing RXRα-TEV, dose-dependent luciferase signals were observed (Fig. 2b
). Once activated, nuclear hormone receptors translocate into the nucleus and interact with nuclear receptor co-activator-1 (NCOA1) to form the transcriptional complex [14Darimont BD, Wagner RL, Apriletti JW, et al. Strucutre and specificity of nuclear receptor-coactivator interactions Genes Dev 1998; 2: 3343-56.]. We tagged retinoid X receptor α (RXRα) with TEV at the C-terminal (RXRα-TEV) and fused a nuclear hormone interaction domain of NCOA1 with the permuted luciferase (NCOA1-pLuc). We expected that active luciferase signals would be generated once RXRα-TEV and NCOA1-pLuc fusion protein make contact in the nucleus. In order to test this, we generated U2OS cells stably expressing NCOA1-pLuc and designated these as U2OS_NCOA1-pLuc cells. When U2OS_NCOA1-pLuc cells were transiently expressing RXRα-TEV, dose-dependent luciferase signals were observed (Fig. 2b ), indicating retinoic acid-induced RXRα-TEV and NCOA1-pLuc complex formation and reconstitution of active luciferase resulted in permuted luciferase cleavage by TEV. Nuclear hormone receptors are classified into multiple subfamilies according their mechanism of action and subcellular distribution in the absence of ligand. RXRα belongs to subfamily II. We next selected estrogen receptor α (ERα), a member of subfamily III and pregnane X receptor (PXR) for further validation of NHR LinkLight assay. When U2OS_NCOA1-pLuc cells transiently expressed ERα-TEV or PXR-TEV, ligand-induced luciferase signals were observed (Fig. 2c
), indicating retinoic acid-induced RXRα-TEV and NCOA1-pLuc complex formation and reconstitution of active luciferase resulted in permuted luciferase cleavage by TEV. Nuclear hormone receptors are classified into multiple subfamilies according their mechanism of action and subcellular distribution in the absence of ligand. RXRα belongs to subfamily II. We next selected estrogen receptor α (ERα), a member of subfamily III and pregnane X receptor (PXR) for further validation of NHR LinkLight assay. When U2OS_NCOA1-pLuc cells transiently expressed ERα-TEV or PXR-TEV, ligand-induced luciferase signals were observed (Fig. 2c and 2d
and 2d ). The results indicated that NHR LinkLight assay technology can be used for broad range of nuclear hormone receptors.
). The results indicated that NHR LinkLight assay technology can be used for broad range of nuclear hormone receptors.
Next, we applied LinkLight assay technology to receptor tyrosine kinases (Fig. 3a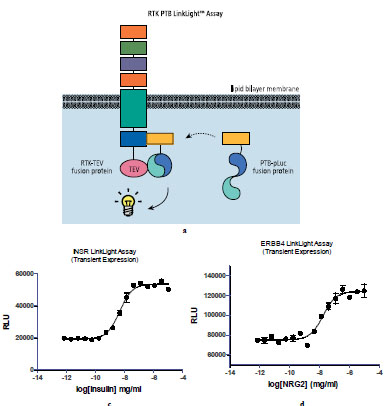 ). Once activated, receptor tyrosine kinases (RTK) recruit Src homology 2 (SH2) and phosphotyrosine binding (PTB) domain containing proteins to initiate signal transduction pathways [15Pawson T. Protein modules and signaling networks Nature 1995; 373: 573-80.]. We used insulin receptor as a model for the RTK LinkLight assay development. We tagged insulin receptor (INSR) with TEV protein and fused a PTB domain with the permuted luciferase. We generated U2OS cells stably expressing PTB-pLuc fusion protein. The cells were then transiently transduced with the INSR-TEV fusion protein and treated with insulin. A dose-dependent EC50 curve was generated (Fig. 3b
). Once activated, receptor tyrosine kinases (RTK) recruit Src homology 2 (SH2) and phosphotyrosine binding (PTB) domain containing proteins to initiate signal transduction pathways [15Pawson T. Protein modules and signaling networks Nature 1995; 373: 573-80.]. We used insulin receptor as a model for the RTK LinkLight assay development. We tagged insulin receptor (INSR) with TEV protein and fused a PTB domain with the permuted luciferase. We generated U2OS cells stably expressing PTB-pLuc fusion protein. The cells were then transiently transduced with the INSR-TEV fusion protein and treated with insulin. A dose-dependent EC50 curve was generated (Fig. 3b ) after adding luciferin, indicating insulin-induced interaction between INSR-TEV and PTB-pLuc fusion proteins and active luciferase generation. We then built other receptor tyrosine kinases in the RTK LinkLight assay. Epidermal growth factor receptor ERBB4 binds ligands such as neuregulin-2 (NRG2) with high affinity and stimulates the receptor tyrosine kinase activity. We constructed ERBB4-TEV expression vectors. Transient expression of ERBB4-TEV in U2OS_PTB-pLuc cells showed ligand-induced signals (Fig. 3c
) after adding luciferin, indicating insulin-induced interaction between INSR-TEV and PTB-pLuc fusion proteins and active luciferase generation. We then built other receptor tyrosine kinases in the RTK LinkLight assay. Epidermal growth factor receptor ERBB4 binds ligands such as neuregulin-2 (NRG2) with high affinity and stimulates the receptor tyrosine kinase activity. We constructed ERBB4-TEV expression vectors. Transient expression of ERBB4-TEV in U2OS_PTB-pLuc cells showed ligand-induced signals (Fig. 3c ).
).
The reconstituted luciferase activity is generally less than 10% of the wild type firefly luciferase activity when equal amounts are expressed in cells. The reduced activity could be due to a less favorable conformation of reconstituted luciferase or partially cleaved permuted luciferase. Since bioluminescent signals are easily detectable, we can read highly sensitive luminescent signals from reconstituted luciferase without over-expressing fusion proteins. Potentially, other commonly used enzymes and fluorescent proteins in reporter gene assays could replace the firefly luciferase by permutation.
CONCLUSION
We report here a novel cell-based assay that can be used to measure protein-protein interactions. The assay method is highly specific, and can be used to assess specific signal pathways. Use of full-length proteins in cell-based assays offers biologically relevant functional readouts, therefore provides better opportunities to identify lead compounds. Given the simplicity, flexibility, sensitivity and the low cost of the assay, we believe the method can be broadly applicable for many other types of protein-protein interactions. Potentially, many new protein-protein interaction drug targets can be explored using LinkLight assay technology offering new drug discovery opportunities. In addition, the LinkLight assay can replace some currently used tedious assay methods and address assay enabling issues for some difficult or intractable drug targets. Furthermore, a new signal pathway assay method can provide opportunities to identify new chemical matters for old targets.
CONFLICTS OF INTEREST
The technology described here was submitted as a patent application and was published in 2009 (WO2009032716; “Identifying molecules that modulate protein-protein interactions using protease activated reporters”).
ACKNOWLEDGEMENTS
Authors thank Jenny Eishingdrelo for graphic design and drawing. Thanks also to Dr. Xiping Bai for providing NRG2.






