- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
A Biochemical/Biophysical Assay Dyad for HTS-Compatible Triaging of Inhibitors of the HIV-1 Nef/Hck SH3 Interaction
Sebastian Breuer1, *, Sheryll Espinola2, Xavier Morelli3, Bruce E Torbett1, Stefan T Arold4, 5, Ingo H Engels2
Abstract
The current treatment regimens for HIV include over 20 anti-retrovirals. However, adverse drug effects and the emergence of drug resistance necessitates the continued improvement of the existing drug classes as well as the development of novel drugs that target as yet therapeutically unexploited viral and cellular pathways. Here we demonstrate a strategy for the discovery of protein-protein interaction inhibitors of the viral pathogenicity factor HIV-1 Nef and its interaction with the host factor SH3. A combination of a time-resolved fluorescence resonance energy resonance energy transfer-based assay and a label-free resonant waveguide grating-based assay was optimized for high-throughput screening formats.
Article Information
Identifiers and Pagination:
Year: 2013Volume: 7
First Page: 16
Last Page: 20
Publisher Id: CCGTM-7-16
DOI: 10.2174/2213988501307010016
Article History:
Received Date: 18/3/2013Revision Received Date: 9/5/2013
Acceptance Date: 20/5/2013
Electronic publication date: 26/7/2013
Collection year: 2013
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Department of Molecular and Experimental Medicine, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA; Tel: +1 858 784 9976; Fax: + 1 858 784 7714; E-mail: breuer@scripps.edu
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 18-3-2013 |
Original Manuscript | A Biochemical/Biophysical Assay Dyad for HTS-Compatible Triaging of Inhibitors of the HIV-1 Nef/Hck SH3 Interaction | |
INTRODUCTION
The negative factor (Nef) is an accessory HIV-1 protein with an extensive cellular interactome and a broad functionality within the HIV replication cycle [1Geyer M, Fackler OT, Peterlin BM. Structure--function relationships in hiv-1 nef EMBO Rep 2001; 2(7 ): 580-., 2Arold ST, Baur AS. Dynamic nef and nef dynamics: How structure could explain the complex activities of this small hiv protein Trends Biochem Sci 2001; 26(6 ): 356-63.]. HIV-1 long-term survivors have been found to harbor viruses with a deficient nef gene, indicating a critical role for Nef in the viral life cycle and in the progression towards AIDS [3Kirchhoff F, Greenough TC, Brettler DB, et al. Brief report.Absence of intact nef sequences in a long-term survivor with nonprogressive hiv-1 infection N Engl J Med 1995; 332(4 ): 228-32., 4Deacon NJ, Tsykin A, Solomon A, et al. Genomic structure of an attenuated quasi species of hiv-1 from a blood transfusion donor and recipients Science Source journal 1995; 270(5238 ): 988-1.]. Nef orchestrates the down-regulation of important surface receptors involved in immune surveillance, such as CD4, MHCI, and MHCII, and activates host cells by triggering signaling pathways involving involving Src family kinases, such as the macrophage-specific hematopoietic cell kinase (Hck) [1Geyer M, Fackler OT, Peterlin BM. Structure--function relationships in hiv-1 nef EMBO Rep 2001; 2(7 ): 580-., 2Arold ST, Baur AS. Dynamic nef and nef dynamics: How structure could explain the complex activities of this small hiv protein Trends Biochem Sci 2001; 26(6 ): 356-63.]. Nef’s functionality is based on numerous protein-protein interactions (PPIs) [2Arold ST, Baur AS. Dynamic nef and nef dynamics: How structure could explain the complex activities of this small hiv protein Trends Biochem Sci 2001; 26(6 ): 356-63., 5Lulf S, Horenkamp FA, Breuer S, Geyer M. Nef surfaces Where to interfere with function. Curr HIV Res 2011; 9(7 ): 543-1.]. The SH3 interaction site on Nef has been identified as a ‘hot spot’ for potential therapeutic intervention due to its highly conserved character [2Arold ST, Baur AS. Dynamic nef and nef dynamics: How structure could explain the complex activities of this small hiv protein Trends Biochem Sci 2001; 26(6 ): 356-63., 5Lulf S, Horenkamp FA, Breuer S, Geyer M. Nef surfaces Where to interfere with function. Curr HIV Res 2011; 9(7 ): 543-1.]. SH3 interacts with a poly-proline type II helix comprising the consensus motif P72xxPxR (HIV-1 NL4-3 nomenclature) and the RT-loop recognition site, a hydrophobic cleft on Nef. The compounds D14, DLC27, and the optimized compound DLC27-14 displayed inhibition of the Nef/SH3 complex formation through targeting of the hydrophobic cleft [6Betzi S, Restouin A, Opi S, et al. Protein protein interaction inhibition (2p2i) combining high throughput and virtual screening: Application to the hiv-1 nef protein Proc Natl Acad Sci USA 2007; 104(49 ): 19256-61., 7Lugari A, Breuer S, Coursindel T, et al. A specific protein disorder catalyzer of hiv-1 nef Bioorg Med Chem 2011; 19(24 ): 7401-6.]. However, all the compounds failed to function in antiviral cell-based assays [6Betzi S, Restouin A, Opi S, et al. Protein protein interaction inhibition (2p2i) combining high throughput and virtual screening: Application to the hiv-1 nef protein Proc Natl Acad Sci USA 2007; 104(49 ): 19256-61., 7Lugari A, Breuer S, Coursindel T, et al. A specific protein disorder catalyzer of hiv-1 nef Bioorg Med Chem 2011; 19(24 ): 7401-6.], thus revealing the need for alternative chemical starting points. To our knowledge, the Nef/SH3 PPI has not been targeted by a biochemical high-throughput screening approach that supports the identification of Nef-specific antivirals.
Here we report on the development and validation of a biochemical time-resolved fluorescence resonance energy transfer (TR-FRET)-based assay for the identification of inhibitors of the Nef/SH3 PPI. The TR-FRET-based assay was paired with an orthogonal biophysical label-free resonant waveguide grating (RWG) assay allowing not only the identification of primary hits that are TR-FRET label-dependent artifacts, but also the confirmation of specifically target-binding compounds.
MATERIAL AND METHODOLOGY
Reagents
All chemicals were purchased from commercial suppliers unless otherwise stated. The HIV-1 His-tagged NefSF2(His-Nef), His-tagged NefSF2 mutant P72xxPxR/AxxAxA and the GST-tagged Hck SH3(GST-SH3) domain were synthesized as recombinant proteins as described previously [8Breuer S, Gerlach H, Kolaric B, et al. Biochemical indication for myristoylation-dependent conformational changes in hiv-1 nef Biochemistry 2006; 45(7 ): 2339-49., 9Breuer S, Schievink SI, Schulte A, et al. Molecular design. functional characterization and structural basis of a protein inhibitor against the hiv-1 pathogenicity factor nef PLoS ONE 2011; 6(5 ): e20033-0.]. The monoclonal Eu-cryptate-conjugate α-GST(α-GST mAb-Eu) and the APC (XL665)-conjugated α-His (α-His mAb-XL) antibodies were purchased from Invitrogen (Carlsbad, CA, USA) and Cisbio-US (Bedford, MA, USA), respectively. The compound D14 was kindly provided by the AIDS reagents program. DLC27 was synthesized as described previously [7Lugari A, Breuer S, Coursindel T, et al. A specific protein disorder catalyzer of hiv-1 nef Bioorg Med Chem 2011; 19(24 ): 7401-6.].
TR-FRET-Based Assay
The TR-FRET-based assay was carried out in white solid-bottom 1536-well plates (Greiner, San Diego, CA, USA). 500 nM His-Nef, 10 nM GST-SH3, 0.5 nM α-GST mAb-Eu and 5 nM α-His mAb-XL were used in PBS in a final detection volume of 5 µl at room temperature. After 2 hrs incubation the fluorescence at 620nm and 665nm was detected using a Pherastar FS plate reader (BMG Labtech, Ortenberg, Germany). All experiments were performed in triplicates.
High Throughput Screening
For the Library of Pharmacologically Active Compounds (LOPAC) (Sigma Aldrich, St Louise, MO, USA) screen individual 50-nl compound aliquots from 1 mM DMSO stocks were transferred using an acoustic dispenser (Labcyte, Sunnyvale, CA, USA) to a well containing the quaternary complex (His-Nef, GST-SH3, α-GST mAb-Eu and α-His mAb-XL) in a 5 µl volume, which was then incubated for 2 h before the TR-FRET-based assay was measured.
RWG-Basedcompetitive Assay
The RWG experiments were carried out in 384-well GA3 aldehyde plates from SRU Biosystems (Woburn, MA) using a SRU BIND® SCREENER reader (SRU Biosystems, Woburn, MA). 5 µM GST-SH3 was coated on a pre-activated biosensor via its primary amines at room temperature. The binding kinetic of 0.15 µM Nef in presence of 50 µM compound or DMSO was measured over time in a final volume of 50 µl, PBS + 1 % (v/v) DMSO buffer.
RWG-Based Direct Binding Assay
A 384-well GA3 aldehyde plate was coated with 5 µM His-Nef (or GST-SH3) and the binding kinetic of 50 µM compound was detected in a final volume of 50 µl in PBS + 1 % (v/v) DMSO buffer using SRU BIND® SCREENER reader (SRU Biosystems, Woburn, MA).
RESULTS
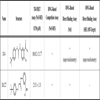
Fig. (1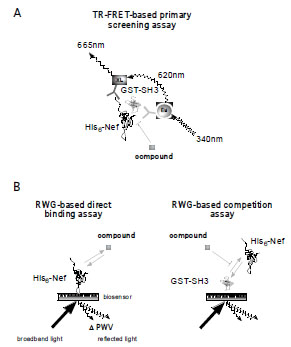 ) shows a schematic of the assay cascade comprising a TR-FRET-based primary screening assay and an orthogonal label-free RWG-based assay. The GST-tagged Hck SH3 domain (GST-SH3) forms a stable complex with the His-tagged Nef protein (His-Nef) in solution (Fig.1A
) shows a schematic of the assay cascade comprising a TR-FRET-based primary screening assay and an orthogonal label-free RWG-based assay. The GST-tagged Hck SH3 domain (GST-SH3) forms a stable complex with the His-tagged Nef protein (His-Nef) in solution (Fig.1A ). The affinity of this interaction was quantified previously and found to have a Kd of 0.25 - 1.5 µM [9Breuer S, Schievink SI, Schulte A, et al. Molecular design. functional characterization and structural basis of a protein inhibitor against the hiv-1 pathogenicity factor nef PLoS ONE 2011; 6(5
): e20033-0., 10Arold S, O'Brien R, Franken P, et al. Rt loop flexibility enhances the specificity of src family sh3 domains for hiv-1 nef Biochemistry 1998; 37(42
): 14683-91.]. The interaction can be detected homogeneously and therefore automation friendly by TR-FRET between a europium(Eu(III))-conjugated anti-GST monoclonal antibody (α-GST mAb-Eu) and an anti-His XL665-labeled mAb (α-His mAb-XL) in a quaternary complex (Fig.1A
). The affinity of this interaction was quantified previously and found to have a Kd of 0.25 - 1.5 µM [9Breuer S, Schievink SI, Schulte A, et al. Molecular design. functional characterization and structural basis of a protein inhibitor against the hiv-1 pathogenicity factor nef PLoS ONE 2011; 6(5
): e20033-0., 10Arold S, O'Brien R, Franken P, et al. Rt loop flexibility enhances the specificity of src family sh3 domains for hiv-1 nef Biochemistry 1998; 37(42
): 14683-91.]. The interaction can be detected homogeneously and therefore automation friendly by TR-FRET between a europium(Eu(III))-conjugated anti-GST monoclonal antibody (α-GST mAb-Eu) and an anti-His XL665-labeled mAb (α-His mAb-XL) in a quaternary complex (Fig.1A ). Upon the Eu (III)-cryptate donor excitation at 340 nm, part of the emission at 620 nm activates the acceptor, allophycocyanin XL665, whose emission is detectable at 665 nm. The longevity of the 620 nm emission allows the time-resolved assessment of the 665/620 nm intensity ratio (as a measure of the binding of SH3 to Nef)100 µs after UV excitation of the quaternary complex. Both the large Stokes shift of the Eu(III)-fluorescence and the time resolution of the measurement eliminate most sources of fluorescence interference, making Eu(III) especially well-suited for high-throughput screening of large and diverse compound libraries [11Mathis G. Htrf(r) technology J Biomol Screen 1999; 4(6
): 309-14., 12Engels IH, Daguia C, Huynh T, et al. A time-resolved fluorescence resonance energy transfer-based assay for den1 peptidase activity Anal Biochem 2009; 390(1
): 85-7.]. In some cases compounds may have inherent fluorescence that confounds the use of screening using a TR-FRET-based assay. The use of the label-free resonant waveguide grating (RWG) technology for evaluation of fluorescent compounds negates this concern (Fig.1B
). Upon the Eu (III)-cryptate donor excitation at 340 nm, part of the emission at 620 nm activates the acceptor, allophycocyanin XL665, whose emission is detectable at 665 nm. The longevity of the 620 nm emission allows the time-resolved assessment of the 665/620 nm intensity ratio (as a measure of the binding of SH3 to Nef)100 µs after UV excitation of the quaternary complex. Both the large Stokes shift of the Eu(III)-fluorescence and the time resolution of the measurement eliminate most sources of fluorescence interference, making Eu(III) especially well-suited for high-throughput screening of large and diverse compound libraries [11Mathis G. Htrf(r) technology J Biomol Screen 1999; 4(6
): 309-14., 12Engels IH, Daguia C, Huynh T, et al. A time-resolved fluorescence resonance energy transfer-based assay for den1 peptidase activity Anal Biochem 2009; 390(1
): 85-7.]. In some cases compounds may have inherent fluorescence that confounds the use of screening using a TR-FRET-based assay. The use of the label-free resonant waveguide grating (RWG) technology for evaluation of fluorescent compounds negates this concern (Fig.1B ). The RWG technology utilizes a photonic crystal biosensor surface that reflects broadband light in a narrow range of wavelength (peak wavelength value, or PWV). Any alteration of the mass of an immobilized biomolecule on the biosensor surface, such as through the binding of a compound, alters the dielectric permittivity of the biosensor material, which causes a shift of the reflected wavelength (ΔPWV) proportional to the change in mass [13Cunningham BT, Li P, Schulz S, et al. Label-free assays on the bind system J Biomol Screen 2004; 9(6
): 481-90.]. Thus, immobilization of the biomolecule on the biosensor surface, followed by the addition of putative chemical or biological binding partners can be monitored kinetically so that one can discriminate binding events on the basis of multiple criteria such as stoichiometry and association kinetics [14Chan LL, Lidstone EA, Finch KE, et al. A method for identifying small-molecule aggregators using photonic crystal biosensor microplates JALA Charlottesv Va 2009; 14(6
): 348-59.].
). The RWG technology utilizes a photonic crystal biosensor surface that reflects broadband light in a narrow range of wavelength (peak wavelength value, or PWV). Any alteration of the mass of an immobilized biomolecule on the biosensor surface, such as through the binding of a compound, alters the dielectric permittivity of the biosensor material, which causes a shift of the reflected wavelength (ΔPWV) proportional to the change in mass [13Cunningham BT, Li P, Schulz S, et al. Label-free assays on the bind system J Biomol Screen 2004; 9(6
): 481-90.]. Thus, immobilization of the biomolecule on the biosensor surface, followed by the addition of putative chemical or biological binding partners can be monitored kinetically so that one can discriminate binding events on the basis of multiple criteria such as stoichiometry and association kinetics [14Chan LL, Lidstone EA, Finch KE, et al. A method for identifying small-molecule aggregators using photonic crystal biosensor microplates JALA Charlottesv Va 2009; 14(6
): 348-59.].
The optimal TR-FRET-based assay conditions were defined in a multi-dimensional approach by varying the protein concentrations of both His-Nef and GST-SH3 and varying the ratios of α-GST mAb-Eu to α-His mAb-XL. Fig. (2A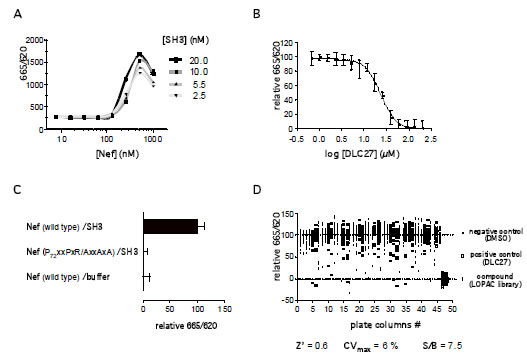 ) shows an example of signal development in response to increasing His-Nef and GST-SH3 concentrations at fixed mAb concentrations. As can be seen, the TR-FRET-based assay signal increased in a His-Nef and GST-SH3 concentration-dependent manner, until at high concentrations free His-Nef competed for the binding of detection antibody and blocked the formation of the quaternary complex, resulting in a decrease of the 665/620 nm ratio. Concentrations of 500 nM His-Nef, 10 nM GST-SH3, 0.5 nM α-GST mAb-Eu and 5 nM α-His mAb-XL in a reaction volume of 5 µl in PBS displayed the best signal to background (S/B = 7.5) values with an adequate signal robustness (coefficient of variations < 10%). These assay parameters tolerated dimethyl sulfoxide (DMSO) concentrations of up to 2 % (v/v), and the signal was stable in a time frame of t = 60 – 180 min after antibody addition (data not shown).
) shows an example of signal development in response to increasing His-Nef and GST-SH3 concentrations at fixed mAb concentrations. As can be seen, the TR-FRET-based assay signal increased in a His-Nef and GST-SH3 concentration-dependent manner, until at high concentrations free His-Nef competed for the binding of detection antibody and blocked the formation of the quaternary complex, resulting in a decrease of the 665/620 nm ratio. Concentrations of 500 nM His-Nef, 10 nM GST-SH3, 0.5 nM α-GST mAb-Eu and 5 nM α-His mAb-XL in a reaction volume of 5 µl in PBS displayed the best signal to background (S/B = 7.5) values with an adequate signal robustness (coefficient of variations < 10%). These assay parameters tolerated dimethyl sulfoxide (DMSO) concentrations of up to 2 % (v/v), and the signal was stable in a time frame of t = 60 – 180 min after antibody addition (data not shown).
Based on the optimized assay conditions the potency of a known inhibitor of the Nef/SH3 interaction, DLC27, was evaluated (Fig.2B ) [6Betzi S, Restouin A, Opi S, et al. Protein protein interaction inhibition (2p2i) combining high throughput and virtual screening: Application to the hiv-1 nef protein Proc Natl Acad Sci USA 2007; 104(49
): 19256-61.]. A DLC27 concentration series was evaluated and the IC50 calculated with a four-parameter logistic model from GraphPad Prism 5.0. The IC50 of 23.51 ± 1.31 (mean ± s.d.) µM correlated with previously published data [6Betzi S, Restouin A, Opi S, et al. Protein protein interaction inhibition (2p2i) combining high throughput and virtual screening: Application to the hiv-1 nef protein Proc Natl Acad Sci USA 2007; 104(49
): 19256-61., 7Lugari A, Breuer S, Coursindel T, et al. A specific protein disorder catalyzer of hiv-1 nef Bioorg Med Chem 2011; 19(24
): 7401-6.]. As a second proof of concept, we tested a His-Nef mutant lacking the canonical PxxPxR motif: it did not show a TR-FRET-based assay signal (Fig.2C
) [6Betzi S, Restouin A, Opi S, et al. Protein protein interaction inhibition (2p2i) combining high throughput and virtual screening: Application to the hiv-1 nef protein Proc Natl Acad Sci USA 2007; 104(49
): 19256-61.]. A DLC27 concentration series was evaluated and the IC50 calculated with a four-parameter logistic model from GraphPad Prism 5.0. The IC50 of 23.51 ± 1.31 (mean ± s.d.) µM correlated with previously published data [6Betzi S, Restouin A, Opi S, et al. Protein protein interaction inhibition (2p2i) combining high throughput and virtual screening: Application to the hiv-1 nef protein Proc Natl Acad Sci USA 2007; 104(49
): 19256-61., 7Lugari A, Breuer S, Coursindel T, et al. A specific protein disorder catalyzer of hiv-1 nef Bioorg Med Chem 2011; 19(24
): 7401-6.]. As a second proof of concept, we tested a His-Nef mutant lacking the canonical PxxPxR motif: it did not show a TR-FRET-based assay signal (Fig.2C ). The feasibility of miniaturization and automation of the TR-FRET-based assay for HTS purposes was examined by testing a 1536-well compound plate from the Library of Pharmacologically Active Compounds (LOPAC, Sigma-Aldrich, St. Louis, MO, USA) (Fig.2D
). The feasibility of miniaturization and automation of the TR-FRET-based assay for HTS purposes was examined by testing a 1536-well compound plate from the Library of Pharmacologically Active Compounds (LOPAC, Sigma-Aldrich, St. Louis, MO, USA) (Fig.2D ). Individual 50-nl compound aliquots from 1 mM DMSO stocks were transferred using an acoustic dispenser (Labcyte, Sunnyvale, CA, USA) to a well containing the quaternary complex in a 5 µl volume, which was then incubated for 2 h before the TR-FRET-based assay was measured. The Z’ factor [15Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays J Biomol Screen 1999; 4(2
): 67-73.] of this representative test compound plate was calculated to be 0.6. Among the LOPAC compounds, we also placed our positive control DLC27 and D14 as test samples. DLC27 and D14 had been identified as Nef-binding small molecules and inhibitors of the Nef/SH3 protein-protein interaction [6Betzi S, Restouin A, Opi S, et al. Protein protein interaction inhibition (2p2i) combining high throughput and virtual screening: Application to the hiv-1 nef protein Proc Natl Acad Sci USA 2007; 104(49
): 19256-61.]. Research on DLC27 has continued in a hit-to-lead study, while D14 was not followed up for further antiviral testing [7Lugari A, Breuer S, Coursindel T, et al. A specific protein disorder catalyzer of hiv-1 nef Bioorg Med Chem 2011; 19(24
): 7401-6.]. Both compounds could be identified as hits and be confirmed in IC50 experiments (Table 1). The robustness and dynamic range of the TR-FRET-based assay allows for both an automated ultra-HTS campaign and ranking of primary hits according to their IC50 values.
). Individual 50-nl compound aliquots from 1 mM DMSO stocks were transferred using an acoustic dispenser (Labcyte, Sunnyvale, CA, USA) to a well containing the quaternary complex in a 5 µl volume, which was then incubated for 2 h before the TR-FRET-based assay was measured. The Z’ factor [15Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays J Biomol Screen 1999; 4(2
): 67-73.] of this representative test compound plate was calculated to be 0.6. Among the LOPAC compounds, we also placed our positive control DLC27 and D14 as test samples. DLC27 and D14 had been identified as Nef-binding small molecules and inhibitors of the Nef/SH3 protein-protein interaction [6Betzi S, Restouin A, Opi S, et al. Protein protein interaction inhibition (2p2i) combining high throughput and virtual screening: Application to the hiv-1 nef protein Proc Natl Acad Sci USA 2007; 104(49
): 19256-61.]. Research on DLC27 has continued in a hit-to-lead study, while D14 was not followed up for further antiviral testing [7Lugari A, Breuer S, Coursindel T, et al. A specific protein disorder catalyzer of hiv-1 nef Bioorg Med Chem 2011; 19(24
): 7401-6.]. Both compounds could be identified as hits and be confirmed in IC50 experiments (Table 1). The robustness and dynamic range of the TR-FRET-based assay allows for both an automated ultra-HTS campaign and ranking of primary hits according to their IC50 values.
To exclude compound artifacts due to fluorescence interference or inhibition of the Nef/SH3 interaction by the virtue of colloidal compound aggregate formation [16McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening J Med Chem 2002; 45(8
): 1712-22.], we paired the primary TR-FRET-based assay, in an orthogonal approach, with a label-free RWG-based assay format using a 384-well plate setup (Fig.1B ). Using the tool compounds DLC27 and D14 for follow up studies, we employed RWG-based competition assays as well as RWG-based direct binding assays to evaluate hits regarding their protein-protein interaction inhibition and direct target binding properties [17Geschwindner S, Carlsson JF, Knecht W. Application of optical biosensors in small-molecule screening activities Sensors (Basel) 2012; 12(4
): 4311-23.]. Fig. (3A
). Using the tool compounds DLC27 and D14 for follow up studies, we employed RWG-based competition assays as well as RWG-based direct binding assays to evaluate hits regarding their protein-protein interaction inhibition and direct target binding properties [17Geschwindner S, Carlsson JF, Knecht W. Application of optical biosensors in small-molecule screening activities Sensors (Basel) 2012; 12(4
): 4311-23.]. Fig. (3A ) shows the binding kinetics of His-Nef to GST-SH3 immobilized on an aldehyde-coated biosensor plate in the presence of 50 µM DLC27 or a control (DMSO). Only the control sample shows the expected shift of the reflected wavelength (ΔPWV), confirming that DLC27 disrupts the interaction of Nef and SH3. To further investigate the binding specificity of compounds to the protein target, we immobilized the viral Nef protein or the human SH3 domain on the biosensor surface and then treated each with DLC27 or D14 (Fig.3B
) shows the binding kinetics of His-Nef to GST-SH3 immobilized on an aldehyde-coated biosensor plate in the presence of 50 µM DLC27 or a control (DMSO). Only the control sample shows the expected shift of the reflected wavelength (ΔPWV), confirming that DLC27 disrupts the interaction of Nef and SH3. To further investigate the binding specificity of compounds to the protein target, we immobilized the viral Nef protein or the human SH3 domain on the biosensor surface and then treated each with DLC27 or D14 (Fig.3B and C
and C ). The real-time binding kinetics of DLC27 or D14 to immobilized His-Nef or GST-SH3 demonstrated that DLC27 selectively bound to Nef, whereas D14 produced a positive PWV shift with both His-Nef and GST-SH3 (Fig.3B
). The real-time binding kinetics of DLC27 or D14 to immobilized His-Nef or GST-SH3 demonstrated that DLC27 selectively bound to Nef, whereas D14 produced a positive PWV shift with both His-Nef and GST-SH3 (Fig.3B , C
, C and Table 1). These findings could indicate that D14 shares a binding site between His-Nef and GST-SH3. However, given that the magnitude of the PWV shifts caused by D14 exceeded what can be expected from either the molecular mass or the stoichiometry of compounds, it appears that D14 is a promiscuous compound that disrupts Nef/SH3 interaction.
and Table 1). These findings could indicate that D14 shares a binding site between His-Nef and GST-SH3. However, given that the magnitude of the PWV shifts caused by D14 exceeded what can be expected from either the molecular mass or the stoichiometry of compounds, it appears that D14 is a promiscuous compound that disrupts Nef/SH3 interaction.
DISCUSSION
In summary, we have developed a robust and HTS-compatible assay comprising a primary TR-FRET-based assay for identifying inhibitors of Nef/SH3 complex formation. Taking advantage of the emergence of HTS-amenable biophysical methodologies, we combined the TR-FRET-based assay with an orthogonal biophysical label-free RWG-based assay, the HTS resolving power is extended, by allowing screening of fluorescent compounds, as well as the identification of Nef/SH3 promiscuous binders. In combining these two distinct methodologies into a high-throughput assay cascade, novel Nef/SH3 inhibitors can be rapidly selected and ranked from large chemical libraries by their inhibitory activity and specific binding early in a drug discovery campaign (Table1).
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
ACKNOWLEDGEMENTS
We gratefully acknowledge the AIDS reagents program for supplying the study with D14 and Dr. Isabelle Parrot for providing DLC27. We thank Walter Pagel for critical reading of the manuscript. The studies were supported by CHRP-F09-SRI-205 (S.B.) and 5P01 GM083658 (BET), 5 R01 and HL091219 (BET). This is publication MEM is #21639 from The Scripps Research Institute.