- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Ornithology Journal
(Discontinued)
ISSN: 1874-4532 ― Volume 13, 2020
Major Histocompatibility Complex Allele Persistence in Eurasia and America in the Genus Carduelis (Spinus) During Million Years
Antonio Arnaiz-Villena2, *, Valentín Ruiz-del-Valle2, Ester Muñiz2, Jose Palacio-Gruber2, Cristina Campos2, Eduardo Gómez-Casado1, Jose Manuel Martín Villa2, Ignacio Serrano-Vela2
Abstract
Introduction:
Genus Carduelis (Fringillidae family) includes goldfinches, siskins, redpolls, greenfinches and crossbills. Many of the species classified within this genus and other related genera have been grouped by using molecular systematics and the mitochondrial cytochrome b (mt cyt b) gene. According to this, the Eurasian siskin (C. spinus) is the only one extant direct ancestor of several North American finches; North American / South American radiations may have been originated by Eurasian siskin (or extinct relative). In the present work, we aim to perform a study of transpecies and transcontinental analyses of MHC (Major Histocompatibility Complex) Class I alleles in several genus Carduelis / Spinus species in order to draw evolutionary conclusions in several wild bird species belonging to the genus Carduelis / Spinus.
Materials and Methods:
Blood was taken from worldwide wild bird species. Passerine phylogeny was done after analysing mtDNA with Maximun Likelihood and Bayesian dendrograms. Major histocompatibility complex alleles were obtained by standard DNA cloning and sequencing.
Results:
We found two matches between MHC-I DNA alleles from different South American siskins at DNA level. Also, it was observed that the Eurasian siskin shares a protein with pine siskin and another with three South American siskins. Eight South American siskins species also share the same MHC protein. In addition, studied songbirds MHC class I intron 2 is longer than that of Gallus gallus.
Conclusion:
We have drawn the following conclusions: 1) We present the first direct evidence that “Minimal Essential MHC” does not exist for birds; one of its main definition characters, i.e.: small intron size does not hold for songbirds. 2) We also report that MHC genes transpecies evolution exist in birds by showing also for the first time that worldwide bird species keep the same MHC protein and DNA alleles. 3) New evidences on MHC alleles conservation from Eurasian Carduelis spinus (most ancient) to South American siskins (most recent) during million years support that Eurasian siskin is the parental species for American Genus Carduelis (Spinus) species. It is uncertain whether Eurasian siskin (or extant relative) had initially an Holoartic distribution, including America.
Article Information
Identifiers and Pagination:
Year: 2017Volume: 10
First Page: 92
Last Page: 104
Publisher Id: TOOENIJ-10-92
DOI: 10.2174/1874453201710010092
Article History:
Received Date: 20/07/2016Revision Received Date: 15/09/2017
Acceptance Date: 20/09/2017
Electronic publication date: 31/10/2017
Collection year: 2017
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: (https://creativecommons.org/licenses/by/4.0/legalcode). This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
* Address correspondence to this author at the Departamento de Immunología, Facultad de Medicina, Universidad Complutense, Pabellón 5, planta 4. Avda. Complutense s/n, 28040 Madrid, Spain; Tel: +34913941642, +34 606993161; E-mail: arnaizantonio@gmail.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 20-07-2016 |
Original Manuscript | Major Histocompatibility Complex Allele Persistence in Eurasia and America in the Genus Carduelis (Spinus) During Million Years | |
1. INTRODUCTION
Genus Carduelis / Spinus includes goldfinches, siskins, redpolls, greenfinches and crossbills [1Arnaiz-Villena A, Guillén J, Ruiz-del-Valle V, et al. Phylogeography of crossbills, bullfinches, grosbeaks, and rosefinches. Cell Mol Life Sci 2001; 58(8): 1159-66.
[http://dx.doi.org/10.1007/PL00000930] [PMID: 11529508] ]. It is comprised within the Fringillidae family of birds together with canaries, many sparrows, bramblings and chaffinches. Most of them are familiar to birdwatchers and urban and country people [2Armani GC. Guide des Passereaux Granivores 1983.-4Clement P, Harris A, Davis J. Finches and Sparrows 1999.]. Many of the species classified within genus Carduelis and other related genera have been grouped by using molecular systematics and the mitochondrial cytochrome b (mt cyt b) gene sequence [5Arnaiz-Villena A, Alvarez-Tejado M, Ruiz-del-Valle V, et al. Rapid radiation of canaries (Genus Serinus). Mol Biol Evol 1999; 16: 2-11.
[http://dx.doi.org/10.1093/oxfordjournals.molbev.a026034] -8Arnaiz-Villena A, Alvarez-Tejado M, Ruíz-del-Valle V, et al. Phylogeny and rapid northern and southern hemisphere speciation of goldfinches during the Miocene and Pliocene epochs. Cell Mol Life Sci 1998; 54(9): 1031-41.
[http://dx.doi.org/10.1007/s000180050230] [PMID: 9791543] ]. According to this, the Eurasian siskin (C. spinus) appeared on Earth in the Pliocene Epoch about 5 million years ago [1Arnaiz-Villena A, Guillén J, Ruiz-del-Valle V, et al. Phylogeography of crossbills, bullfinches, grosbeaks, and rosefinches. Cell Mol Life Sci 2001; 58(8): 1159-66.
[http://dx.doi.org/10.1007/PL00000930] [PMID: 11529508] , 8Arnaiz-Villena A, Alvarez-Tejado M, Ruíz-del-Valle V, et al. Phylogeny and rapid northern and southern hemisphere speciation of goldfinches during the Miocene and Pliocene epochs. Cell Mol Life Sci 1998; 54(9): 1031-41.
[http://dx.doi.org/10.1007/s000180050230] [PMID: 9791543] -10Arnaiz-Villena A, Ruiz-del-Valle V, Moscoso J, Serrano-Vela JI, Zamora J. mtDNA phylogeny of North American Carduelis pinus group. Ardeola 2007; 54: 1-14.]. It is the one extant direct ancestor of several North American finches, which appeared around 2 million years ago [8Arnaiz-Villena A, Alvarez-Tejado M, Ruíz-del-Valle V, et al. Phylogeny and rapid northern and southern hemisphere speciation of goldfinches during the Miocene and Pliocene epochs. Cell Mol Life Sci 1998; 54(9): 1031-41.
[http://dx.doi.org/10.1007/s000180050230] [PMID: 9791543] , 10Arnaiz-Villena A, Ruiz-del-Valle V, Moscoso J, Serrano-Vela JI, Zamora J. mtDNA phylogeny of North American Carduelis pinus group. Ardeola 2007; 54: 1-14.-12Arnaiz-Villena A, Ruiz-del-Valle V, Reguera R, Gomez-Prieto P, Serrano-Vela JI. What might have been the ancestor of New World siskins? Open Ornithol J 2008; 1: 46-7.
[http://dx.doi.org/10.2174/1874453200801010046] ]: C. dominicensis, the Antillean siskin from the Caribbean high peaks of La Hispaniola Island; C. pinus, the pine siskin from North America; and C. atriceps, the Black-capped siskin from Guatemalan - Mexican altiplano [10Arnaiz-Villena A, Ruiz-del-Valle V, Moscoso J, Serrano-Vela JI, Zamora J. mtDNA phylogeny of North American Carduelis pinus group. Ardeola 2007; 54: 1-14., 11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
[http://dx.doi.org/10.2174/1874453201205010073] ]. In addition, North American C. tristis and C. notata (also in South America) radiations may have been originated by Eurasian siskin (C. spinus) entrance to America [7Arnaiz-Villena A, Gomez-Prieto P, Ruiz-del-Valle V. Phylogeography of finches and sparrows 2009; 1-54., 8Arnaiz-Villena A, Alvarez-Tejado M, Ruíz-del-Valle V, et al. Phylogeny and rapid northern and southern hemisphere speciation of goldfinches during the Miocene and Pliocene epochs. Cell Mol Life Sci 1998; 54(9): 1031-41.
[http://dx.doi.org/10.1007/s000180050230] [PMID: 9791543] , 11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
[http://dx.doi.org/10.2174/1874453201205010073] , 12Arnaiz-Villena A, Ruiz-del-Valle V, Reguera R, Gomez-Prieto P, Serrano-Vela JI. What might have been the ancestor of New World siskins? Open Ornithol J 2008; 1: 46-7.
[http://dx.doi.org/10.2174/1874453200801010046] ].
On the other hand, Major Histocompatibility Complex (MHC) is the most polymorphic loci in humans and studied vertebrates and its molecules present antigenic peptides to clonotypic T-cell receptors in order to start the immune response [13Klein J. Natural history of the Major Histocompatibility Complex 1986.]. Classical MHC-I molecules are expressed on all nucleated cells and inhabit receptor structures that bind short peptides (antigens) derived from intracellular pathogens (like viruses) as well as peptides of individual’s own body. After an antigen has been bound, the MHC-antigen complex is transported to the cell surface where it is recognized by CD8+ T-cells. When the presented antigen is from a pathogen, CD8+ T-cell becomes activated and the infected host cell is killed [14Murphy K, Travers P, Walport M. Janeway’s immunobiology 2008.]. Non-classical MHC-I molecules (class-Ib) have a similar structure to classical class I molecules but they are less polymorphic and are not expressed to the same extent [15Gomez-Prieto P, Parga-Lozano C, Rey D, Moreno E, Arnaiz-Villena A. HLA-G, -F and -E: polymorphism, function and evolution 2010; 159-74.]. These class – Ib proteins evolve rapidly and are quite different in primary sequence among different vertebrate species [15Gomez-Prieto P, Parga-Lozano C, Rey D, Moreno E, Arnaiz-Villena A. HLA-G, -F and -E: polymorphism, function and evolution 2010; 159-74., 16Donadi EA, Castelli EC, Arnaiz-Villena A, et al. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci 2011; 68(3): 369-95.
[http://dx.doi.org/10.1007/s00018-010-0580-7] [PMID: 21107637] ]. In the last years, non-classical class I MHC (HLA) genes are currently being studied: its function is immune modulation (in order to avoid autoimmunity) and immune suppression (in order to maintain mother/father proteins compatibility) and prevent fetus infection as a transplant (for an extensive review see [16Donadi EA, Castelli EC, Arnaiz-Villena A, et al. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci 2011; 68(3): 369-95.
[http://dx.doi.org/10.1007/s00018-010-0580-7] [PMID: 21107637] ]). MHC evolution is proposed to be under several kinds of selection: a) balancing selection, i.e.: many different alleles are maintained in the population because they may be beneficial [17Graur D, Li WH. Fundamentals of molecular evolution 2nd ed. 1999.], thus heterozygosis is favoured in order to maximize the range of antigens that can be recognized [18Lewontin RC, Hubby JL. A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics 1966; 54(2): 595-609.
[PMID: 5968643] -20Black FL, Salzano FM. Evidence for heterosis in the HLA system. Am J Hum Genet 1981; 33(6): 894-9.
[PMID: 7325154] ]; b) frequency-dependent selection. Rare alleles would be privileged in the prevention of rare pathogens appearance, which may unexpectedly enter into a population [13Klein J. Natural history of the Major Histocompatibility Complex 1986., 21Bodmer WF, Bodmer JG. Evolution and function of the HLA system. Br Med Bull 1978; 34(3): 309-16.
[http://dx.doi.org/10.1093/oxfordjournals.bmb.a071518] [PMID: 363227] ]; c) a transpecific evolution; this has been observed also in humans and primates [22Klein J, Sato A, Nagl S. O’hUigin C. Molecular trans-species polymorphism. Annu Rev Ecol Syst 1998; 29: 1-21.
[http://dx.doi.org/10.1146/annurev.ecolsys.29.1.1] , 23Arnaiz-Villena A, Enriquez-de-Salamanca M, Palacio-Grüber J, et al. Characterisation and functional implications of the two new HLA-G alleles found in Amerindian and Caribbean populations. Hum Immunol 2016; 77(9): 812-6.
[http://dx.doi.org/10.1016/j.humimm.2016.01.006] [PMID: 26796363] ]. Some specific alleles are markedly favoured and are preserved through speciation, so they can be found in different, genetically related species.
In this high variability context which may include non-classical class I molecules [16Donadi EA, Castelli EC, Arnaiz-Villena A, et al. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci 2011; 68(3): 369-95.
[http://dx.doi.org/10.1007/s00018-010-0580-7] [PMID: 21107637] , 23Arnaiz-Villena A, Enriquez-de-Salamanca M, Palacio-Grüber J, et al. Characterisation and functional implications of the two new HLA-G alleles found in Amerindian and Caribbean populations. Hum Immunol 2016; 77(9): 812-6.
[http://dx.doi.org/10.1016/j.humimm.2016.01.006] [PMID: 26796363] ], it is relevant that some proteins of a locus or loci may remain identical in quite distant species which have diverged in a relatively long time lag [22Klein J, Sato A, Nagl S. O’hUigin C. Molecular trans-species polymorphism. Annu Rev Ecol Syst 1998; 29: 1-21.
[http://dx.doi.org/10.1146/annurev.ecolsys.29.1.1] ]. This has been observed in vertebrate Major Histocompatibility Complex [24Arnaiz-Villena A, Morales P, Gomez-Casado E, et al. Evolution of MHC-G in primates: A different kind of molecule for each group of species. J Reprod Immunol 1999; 43(2): 111-25.
[http://dx.doi.org/10.1016/S0165-0378(99)00026-1] [PMID: 10479048] ] and Plant Histocompatibility System [18Lewontin RC, Hubby JL. A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics 1966; 54(2): 595-609.
[PMID: 5968643] ]. It usually occurs when a strong positive directional evolutive selection is acting on molecules. In this respect, primate species sometimes may share the same MHC class I molecules [25Suárez B, Morales P, Castro MJ, et al. Mhc-E polymorphism in Pongidae primates: The same allele is found in two different species. Tissue Antigens 1997; 50(6): 695-8.
[http://dx.doi.org/10.1111/j.1399-0039.1997.tb02938.x] [PMID: 9458133] ]. Also, molecular analyses based on MHC class I molecules in songbirds, particularly species from genus Serinus and Carduelis, reveal that these species have two different residues in positions 10 and 96 of the molecule when they are compared with different vertebrate species [26Arnaiz-Villena A, Ruiz-del-Valle V, Reche P, et al. Songbirds conserved sites and intron size of MHC Class I molecules reveal a unique evolution in vertebrates. Open Ornithol J 2010; 3: 156-65.
[http://dx.doi.org/10.2174/1874453201003010156] ].
In the present study, we describe new MHC songbirds molecules in genus Carduelis species which have radiated within million years time lag in distant Earth areas and continents. Thus, we perform a study among species, which thrive in transcontinental habitats of MHC class I alleles in several genus Carduelis species in order to draw evolutionary conclusions in these freely occurring wild vertebrate species belonging to the same genus (Carduelis).
2. MATERIALS AND METHODS
2.1. Sampling
Blood was collected from 36 individuals of wild bird Carduelis species (Table 1). Distribution and cytochrome b GenBank sequence accession numbers are given in Table 1. Blood was obtained from wild birds in their natural thriving areas, and kept at 4°C with an ethylenediaminetetraacetic acid (EDTA) solution until use [26Arnaiz-Villena A, Ruiz-del-Valle V, Reche P, et al. Songbirds conserved sites and intron size of MHC Class I molecules reveal a unique evolution in vertebrates. Open Ornithol J 2010; 3: 156-65.
[http://dx.doi.org/10.2174/1874453201003010156] ].
2.2. DNA Extraction
DNA was isolated from whole blood with an automatic DNA extracting device (Nucleic Acid Extraction System. QuickGene-810, FUJIFILM) after treating samples with a commercial kit (QuickGene Whole Blood Extraction Kit S, FUJIFILM). DNA concentration was measured with a spectrophotometer (ND-1000, NANODROP), and adjusted to about 100 ng/μl. Finally, samples were stored at -20°C.
2.3. MHC and mtDNA Cyt b Amplification, Cloning and Sequencing
Sequences of the MHC molecule most variable region (exon 2, intron 2, exon 3) were amplified by the polymerase chain reaction (PCR) method [27Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol 1987; 155: 335-50.
[http://dx.doi.org/10.1016/0076-6879(87)55023-6] [PMID: 3431465] ] in 42 cycles (10 s at 95°C, 30 s at 65°C, 60 s at 72°C) using an EPPENDORF thermocycler and AmpliTaq DNA Polymerase (APPLIED BIOSYSTEMS) [28Saiki RK, Scharf S, Faloona F, et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 1985; 230(4732): 1350-4.
[http://dx.doi.org/10.1126/science.2999980] [PMID: 2999980] ]. Primers were used as described in [26Arnaiz-Villena A, Ruiz-del-Valle V, Reche P, et al. Songbirds conserved sites and intron size of MHC Class I molecules reveal a unique evolution in vertebrates. Open Ornithol J 2010; 3: 156-65.
[http://dx.doi.org/10.2174/1874453201003010156] ]. Fragments of about 850 base pairs were obtained; some of them were purified by electrophoresis in a 2% agarose gel in order to verify the amplification process. Sequencing was performed using the Sanger method [29Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 1977; 74(12): 5463-7.
[http://dx.doi.org/10.1073/pnas.74.12.5463] [PMID: 271968] ], using the same primers as for amplification, plus an internal primer: 5'- GGATTGATGTGGCTCCAAGG-3'. Ambiguities due to heterocygosity were solved by cloning in competent Escherichia coli cells. An average of 12 different cloned sequences per individual were obtained. Amplification and sequencing of cyt b gene 924 base pairs (bp) was performed as previously described [30Zamora J, Lowy E, Ruiz-del-Valle V, et al. Rhodopechys obsoleta (desert finch): A pale ancestor of greenfinches (Carduelis spp.) according to molecular phylogeny. J Ornithol 2006; 147: 448-56.
[http://dx.doi.org/10.1007/s10336-005-0036-2] ].
2.4. Phylogenetic Analyses
MEGA 5.0 software [31Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28(10): 2731-9.
[http://dx.doi.org/10.1093/molbev/msr121] [PMID: 21546353] ] was used to align MHC sequences and translate DNA sequences into protein, for each individual and clone. Different alleles were identified and characterized manually.
Regarding to cyt-b gene, sequences were further analyzed with MEGA 5.0 as described [31Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28(10): 2731-9.
[http://dx.doi.org/10.1093/molbev/msr121] [PMID: 21546353] ]. Phylogenetic dendrograms were obtained using Maximum Likelihood (ML) methodology [32Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol 1981; 17(6): 368-76.
[http://dx.doi.org/10.1007/BF01734359] [PMID: 7288891] ] with PAUP* v. 4.0b10 program [33Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (* and other methods) version 4 2002.] and Bayesian Inference (BI) methodology using MrBayes program [34Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001; 17(8): 754-5.
[http://dx.doi.org/10.1093/bioinformatics/17.8.754] [PMID: 11524383] , 35Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003; 19(12): 1572-4.
[http://dx.doi.org/10.1093/bioinformatics/btg180] [PMID: 12912839] ]. Model test v. 3.7 [36Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998; 14(9): 817-8.
[http://dx.doi.org/10.1093/bioinformatics/14.9.817] [PMID: 9918953] ] was used to find out a DNA substitution model that fits the data best. Also, best model was used prior to both ML and BI analyses. Linearized ML dendrograms were obtained with PAUP* v. 4.0b10 [33Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (* and other methods) version 4 2002.] with the estimated branch length [37Thorne JL, Kishino H, Painter IS. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol 1998; 15(12): 1647-57.
[http://dx.doi.org/10.1093/oxfordjournals.molbev.a025892] [PMID: 9866200] ] which assumes that the rates among the evolutionary lineages may not be constant. Tree calculation strategy consisted of a heuristic search with NNI (Nearing Neighbour Interchange) swapping algorithm. Robustness of nodes was assessed by 1000 bootstrap replicates in the ML analyses. The parameters rates defining the model of evolution were allowed to change in the BI analysis after each generation in order to increase the likelihood of resulting trees. Therefore, none of the parameters were a priori fixed. In BI analyses, two independent runs (with one cold and three heated chains each) were performed along with 5 million generations. Trees were sampled every 100 generations and the first 12,500 samples were discarded as ‘burn-in’. Split frequencies average standard deviation approached to zero being around 0.01 at the end of the analysis. Posterior probability values (ppv) indicate the robustness of the nodes in the BI. In Phylogenetic analyses chaffinch (Fringilla coelebs) (family Fringillidae, subfamily Fringillinae), was used as outgroup.
3. RESULTS
3.1. Phylogenetic Trees and Age of Appearance on Earth
The peopling of America continent by genus Carduelis species could be carried out by three rapid radiations according to extant present day species: A Mesoamerican goldfinch radiation, a North American siskin radiation and a South American siskin radiation [11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
[http://dx.doi.org/10.2174/1874453201205010073] ]. In this work, species from South American and North American groups have been studied. The age of appearance on Earth of the extant ancestor of North American group, Eurasian siskin (C. spinus) is approximately 5 million years ago [8Arnaiz-Villena A, Alvarez-Tejado M, Ruíz-del-Valle V, et al. Phylogeny and rapid northern and southern hemisphere speciation of goldfinches during the Miocene and Pliocene epochs. Cell Mol Life Sci 1998; 54(9): 1031-41.
[http://dx.doi.org/10.1007/s000180050230] [PMID: 9791543] , 11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
[http://dx.doi.org/10.2174/1874453201205010073] , 12Arnaiz-Villena A, Ruiz-del-Valle V, Reguera R, Gomez-Prieto P, Serrano-Vela JI. What might have been the ancestor of New World siskins? Open Ornithol J 2008; 1: 46-7.
[http://dx.doi.org/10.2174/1874453200801010046] ] (Figs. 1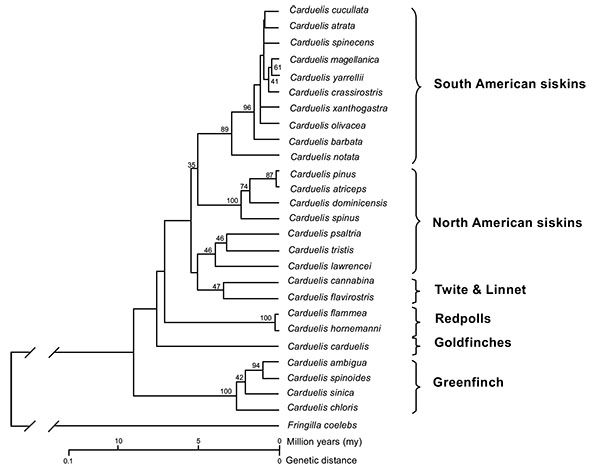 and 2
and 2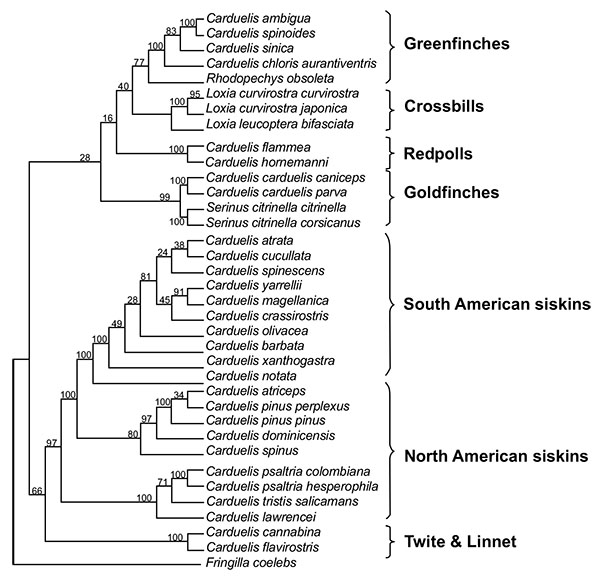 ). It is suggested that this bird passed to America through Beringia/Aleutian Islands [38McLaren IA, Morlan J, Smith P, Gosselin M, Bailey SF. Eurasian siskins in North America - distinguishing females from green-morp pine siskins. American Birds 1989; 43: 1268-74.], since there have been sightings of these birds in areas near these islands. However, it is possible that C. spinus entering to America came also through the East Coast (Greenland, Iceland, Newfoundland): both East and West entering may have possible [11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
). It is suggested that this bird passed to America through Beringia/Aleutian Islands [38McLaren IA, Morlan J, Smith P, Gosselin M, Bailey SF. Eurasian siskins in North America - distinguishing females from green-morp pine siskins. American Birds 1989; 43: 1268-74.], since there have been sightings of these birds in areas near these islands. However, it is possible that C. spinus entering to America came also through the East Coast (Greenland, Iceland, Newfoundland): both East and West entering may have possible [11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
[http://dx.doi.org/10.2174/1874453201205010073] ]. Eurasian siskin (or extinct close relative) evolves to Antillean siskin (C. dominicensis) during the Pliocene Epoch due to a geographical isolation after reaching Antillean area. Pine siskin (C. pinus) seems to be the descendent of Antillean siskin (Figs. 1 and 2
and 2 ). Regarding the South American siskin group, the Black-headed siskin (C. notata) is suggested to be as extant ancestor of this group. South American radiation occurred probably after 3 million years ago, and C. notata or an extinct ancestor passed to South America from Mexican mountains, probably after Isthmus of Panama emerged [11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
). Regarding the South American siskin group, the Black-headed siskin (C. notata) is suggested to be as extant ancestor of this group. South American radiation occurred probably after 3 million years ago, and C. notata or an extinct ancestor passed to South America from Mexican mountains, probably after Isthmus of Panama emerged [11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
[http://dx.doi.org/10.2174/1874453201205010073] ]. This fact favoured the invasion of mesothermal plants from the Rocky Mountains to Andean Spine causing the expansion of this species Carduelis / Spinus genus and triggering the radiation [11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
[http://dx.doi.org/10.2174/1874453201205010073] ].
North American siskin group and South American siskin group are closely related. Both were separated almost 5 million years ago (Fig. 1 ). However, the common precursor of South and North American radiations, namely, the link between Eurasian siskin (C. spinus) and Black-headed siskin (C. notata) is missing, as well as the common ancestor of all three groups (North, Meso and South radiations) [11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
). However, the common precursor of South and North American radiations, namely, the link between Eurasian siskin (C. spinus) and Black-headed siskin (C. notata) is missing, as well as the common ancestor of all three groups (North, Meso and South radiations) [11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
[http://dx.doi.org/10.2174/1874453201205010073] ], unless that C. spinus was the American siskin ancestor and several links are missed [12Arnaiz-Villena A, Ruiz-del-Valle V, Reguera R, Gomez-Prieto P, Serrano-Vela JI. What might have been the ancestor of New World siskins? Open Ornithol J 2008; 1: 46-7.
[http://dx.doi.org/10.2174/1874453200801010046] ].
 |
Fig. (1) Linearized Maximum likelihood dendrogram based on mitochondrial cytochrome b DNA sequences. Fringilla coelebs was chosen as outgroup. |
 |
Fig. (2) Linearized Bayesian dendrogram based on mitochondrial cytochrome b DNA sequences. Fringilla coelebs was chosen as outgroup. |
3.2. New MHC Alleles Found in Genus Carduelis
Once collected all MHC DNA sequences, they were classified according to DNA coding groups of similar or identical proteins. Then, a new provisional nomenclature of alleles was carried out according to the proposal of J. Klein [39Klein J, Bontrop RE, Dawkins RL, et al. Nomenclature for the major histocompatibility complexes of different species: A proposal. Immunogenetics 1990; 31(4): 217-9.
[http://dx.doi.org/10.1007/BF00204890] [PMID: 2329006] ], (Fig. (3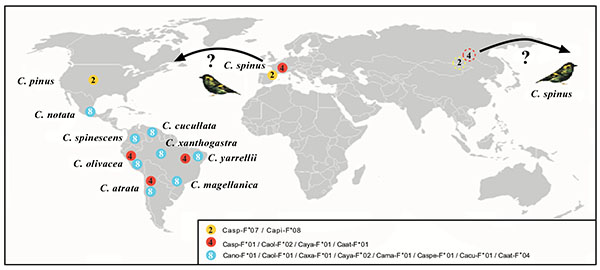 ) footnote). The MHC locus was generically identified with the letter F, referring to chicken class I genes (BF complex), since found alleles have been compared to human and other species class I molecules and found to belong to class I type of MHC molecules [26Arnaiz-Villena A, Ruiz-del-Valle V, Reche P, et al. Songbirds conserved sites and intron size of MHC Class I molecules reveal a unique evolution in vertebrates. Open Ornithol J 2010; 3: 156-65.
) footnote). The MHC locus was generically identified with the letter F, referring to chicken class I genes (BF complex), since found alleles have been compared to human and other species class I molecules and found to belong to class I type of MHC molecules [26Arnaiz-Villena A, Ruiz-del-Valle V, Reche P, et al. Songbirds conserved sites and intron size of MHC Class I molecules reveal a unique evolution in vertebrates. Open Ornithol J 2010; 3: 156-65.
[http://dx.doi.org/10.2174/1874453201003010156] ]. MHC class I molecules often different genus Carduelis species have been studied in the present work. Details of appearance on Earth and habitat of South American siskins are accounted in Table 2 footnote (low case letter).
Eurasian siskin (C. spinus) thrives in Paleartic and Oriental forests and conifer woodland in summer, whereas in winter it is observed in common weedy areas, plantations and gardens [4Clement P, Harris A, Davis J. Finches and Sparrows 1999.]. Eurasian siskin appeared on Earth 5 million years ago: this is observed in the linearized Maximum Likelihood dendrogram (Fig. 1 ). It is the extant ancestor of the North American goldfinch group [1Arnaiz-Villena A, Guillén J, Ruiz-del-Valle V, et al. Phylogeography of crossbills, bullfinches, grosbeaks, and rosefinches. Cell Mol Life Sci 2001; 58(8): 1159-66.
). It is the extant ancestor of the North American goldfinch group [1Arnaiz-Villena A, Guillén J, Ruiz-del-Valle V, et al. Phylogeography of crossbills, bullfinches, grosbeaks, and rosefinches. Cell Mol Life Sci 2001; 58(8): 1159-66.
[http://dx.doi.org/10.1007/PL00000930] [PMID: 11529508] , 7Arnaiz-Villena A, Gomez-Prieto P, Ruiz-del-Valle V. Phylogeography of finches and sparrows 2009; 1-54., 8Arnaiz-Villena A, Alvarez-Tejado M, Ruíz-del-Valle V, et al. Phylogeny and rapid northern and southern hemisphere speciation of goldfinches during the Miocene and Pliocene epochs. Cell Mol Life Sci 1998; 54(9): 1031-41.
[http://dx.doi.org/10.1007/s000180050230] [PMID: 9791543] , 10Arnaiz-Villena A, Ruiz-del-Valle V, Moscoso J, Serrano-Vela JI, Zamora J. mtDNA phylogeny of North American Carduelis pinus group. Ardeola 2007; 54: 1-14., 12Arnaiz-Villena A, Ruiz-del-Valle V, Reguera R, Gomez-Prieto P, Serrano-Vela JI. What might have been the ancestor of New World siskins? Open Ornithol J 2008; 1: 46-7.
[http://dx.doi.org/10.2174/1874453200801010046] ]. In this study, twenty different alleles of C. spinus were found in 14 individuals (Table 2). This species may have been ancestor of all North American siskins [1Arnaiz-Villena A, Guillén J, Ruiz-del-Valle V, et al. Phylogeography of crossbills, bullfinches, grosbeaks, and rosefinches. Cell Mol Life Sci 2001; 58(8): 1159-66.
[http://dx.doi.org/10.1007/PL00000930] [PMID: 11529508] , 7Arnaiz-Villena A, Gomez-Prieto P, Ruiz-del-Valle V. Phylogeography of finches and sparrows 2009; 1-54., 8Arnaiz-Villena A, Alvarez-Tejado M, Ruíz-del-Valle V, et al. Phylogeny and rapid northern and southern hemisphere speciation of goldfinches during the Miocene and Pliocene epochs. Cell Mol Life Sci 1998; 54(9): 1031-41.
[http://dx.doi.org/10.1007/s000180050230] [PMID: 9791543] , 10Arnaiz-Villena A, Ruiz-del-Valle V, Moscoso J, Serrano-Vela JI, Zamora J. mtDNA phylogeny of North American Carduelis pinus group. Ardeola 2007; 54: 1-14., 12Arnaiz-Villena A, Ruiz-del-Valle V, Reguera R, Gomez-Prieto P, Serrano-Vela JI. What might have been the ancestor of New World siskins? Open Ornithol J 2008; 1: 46-7.
[http://dx.doi.org/10.2174/1874453200801010046] ]. Its close relative Pine siskin (C. pinus) inhabits in all North America and goes South to Guatemala. It may be observed in conifer forests, plantations, thickets and shrubs [4Clement P, Harris A, Davis J. Finches and Sparrows 1999.]. This bird is living on Earth since 200,000 years ago, as it is deduced from Fig. (1 ). Eight different alleles of pine siskin were found in 6 individuals in this present study (Table 2).
). Eight different alleles of pine siskin were found in 6 individuals in this present study (Table 2).
DNA alleles were compared with each other (Table 2). Similarities were found: Caat-F*0101 C. atrata allele also appears in C. olivacea (Caol -F*0201) and Caat-F*0401 C. atrata allele is present in other 6 species of South American siskins, such as C. spinescens (Caspe-F*0101), C. xanthogastra (Caxa-F*0101), C. olivacea (Caol-F*0101), C. notata (Cano-F*0101), C. magellanica (Cama-F*0101) and C. cucullata (Cacu-F*0102).
 |
Fig. (3) Map which shows the geographic location of species which share MHC-I proteins. Proteins of different species are named as follows: Carduelis spinus (Casp-F*); Carduelis pinus (Capi-F*); Carduelis notata (Cano-F*); Carduelis spinescens (Caspe-F*); Carduelis olivacea (Caol-F*); Carduelis atrata (Caat-F*); Carduelis magellanica (Cama-F*); Carduelis yarrellii (Caya-F*); Carduelis xanthogastra (Caxa-F*) and Carduelis cucullata (Cacu-F*). Color of the circles indicates the three different proteins found and number inside each circle indicates species that share each protein (Fig. 4  ). ).
|
 |
Fig. (4) Carduelis spinus (left side) and Carduelis atrata (right side). Carduelis spinus appeared on Earth 5 MYA and Carduelis atrata appeared on Earth 0.5-1 MYA [8Arnaiz-Villena A, Alvarez-Tejado M, Ruíz-del-Valle V, et al. Phylogeny and rapid northern and southern hemisphere speciation of goldfinches during the Miocene and Pliocene epochs. Cell Mol Life Sci 1998; 54(9): 1031-41. [http://dx.doi.org/10.1007/s000180050230] [PMID: 9791543] , 10Arnaiz-Villena A, Ruiz-del-Valle V, Moscoso J, Serrano-Vela JI, Zamora J. mtDNA phylogeny of North American Carduelis pinus group. Ardeola 2007; 54: 1-14.-12Arnaiz-Villena A, Ruiz-del-Valle V, Reguera R, Gomez-Prieto P, Serrano-Vela JI. What might have been the ancestor of New World siskins? Open Ornithol J 2008; 1: 46-7. [http://dx.doi.org/10.2174/1874453200801010046] ]. |
When MHC protein alleles are compared, it is observed that C. spinus shared a allele with C. pinus (Casp-F*07 = Capi-F*08) and another one with C. atrata (Casp-F*01 = Caat-F*01) (Fig. 3 ). Furthermore, if South American siskins proteins are considered, it is observed that Casp-F*01 protein is the same one than Caol-F*02 (C. olivacea) and Caya-F*01 (C. yarrellii). Fig. (3
). Furthermore, if South American siskins proteins are considered, it is observed that Casp-F*01 protein is the same one than Caol-F*02 (C. olivacea) and Caya-F*01 (C. yarrellii). Fig. (3 ) shows one the most shared protein (blue) that is present in 8 species of South American siskins; the most worldwide extended protein (red) appears both in Europe and in South America. And finally, there are 3 species which share 2 proteins (C. olivacea, C. atrata y C. yarrellii, blue and red).
) shows one the most shared protein (blue) that is present in 8 species of South American siskins; the most worldwide extended protein (red) appears both in Europe and in South America. And finally, there are 3 species which share 2 proteins (C. olivacea, C. atrata y C. yarrellii, blue and red).
Therefore a total of 2 DNA allele sequences present in 7 different species were found: (C. atrata (Caat-F*0401), C. spinescens (Caspe-F*0101), C. xanthogastra (Caxa-F*0101), C. olivacea (Caol-F*0101), C. notata (Cano-F*0101), C. magellanica (Cama-F*0101), C. cucullata (Cacu-F*0102)) and in 2 (C. atrata (Caat-F*0101) and C. olivacea (Caol -F*0201)) and 3 protein allele sequences are showed in 8 different species (C. notata (Cano-F*01), C. olivacea (Caol-F*01), C. xanthogastra (Caxa-F*01), C. yarrellii (Caya-F*02); C. magellanica (Cama-F*01), C. spinus (Casp-F*01); C. cucullata (Cacu-F*01); C. atrata (Caat-F*04)), 4 (C. spinus (Casp-F*01), C. olivacea (Caol-F*02), C. yarrellii (Caya-F*01), C. atrata (Caat-F*01)) and 2 (C. spinus (Casp-F*07), C. pinus (Capi-F*08)). South American siskins are the species that most sequences have in common, and the Eurasian siskin (C. spinus) shares proteins with the pine siskin (C. pinus) and even South American siskins C. atrata, C. olivacea and C. yarrellii (Fig. 3 ). A preliminary comparison of these DNA sequences with those found in Passer domesticus class I sequences [40Bonneaud C, Sorci G, Morin V, et al. Diversity of Mhc class I and IIB genes in house sparrows (Passer domesticus). Immunogenetics 2004; 55(12): 855-65.
). A preliminary comparison of these DNA sequences with those found in Passer domesticus class I sequences [40Bonneaud C, Sorci G, Morin V, et al. Diversity of Mhc class I and IIB genes in house sparrows (Passer domesticus). Immunogenetics 2004; 55(12): 855-65.
[http://dx.doi.org/10.1007/s00251-004-0648-3] [PMID: 14963619] ] do not show close relatedness with this other species.
4. DISCUSSION
4.1. The Extant Ancestor: Eurasian siskin (C. spinus)
Eurasian siskin migratory behaviour is unpredictable each year: its North to South migrations do not always follows the same longitudinal patterns (it is “irruptive”) [4Clement P, Harris A, Davis J. Finches and Sparrows 1999.]. Nowadays, this bird does not thrive in America, but it lives in easternmost and westernmost Eurasia, being a gap between Central Russia and its easternmost range. It is possible that Eurasian siskin was thriving in Eurasia and also in North America about Pliocene / Pleistocene Epoch limits, approximately 2 million years ago. Later, the Eurasian siskin might have advanced to Caribbean Islands and to Mexican mountains and Guatemalan-Mexican altiplano. About 200,000 years ago, the Eurasian siskin might have given rise to pine siskin (C. pinus) in Mexican sierras and to Antillean siskin (C. dominicensis), nowadays isolated in Hispaniola Island [11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
[http://dx.doi.org/10.2174/1874453201205010073] ,41Lomolino MV, Riddle BR, Brown JH. Biogeography 3rd ed. 2006.]; documented rainfall variations in the Caribbean during the Pleistocene, however, could have also affected distribution of these birds [42Pregill GK, Olson SL. Zoogeography of west indian vertebrates in relation to pleistocene climatic cycles. Annu Rev Ecol Syst 1981; 12: 75-98.
[http://dx.doi.org/10.1146/annurev.es.12.110181.000451] ]. This may be a typical example of adaptive radiation originated by a North to South migration barrier and provincialism that drove evolution to create these new finch species. Last Wisconsin Glaciation ended and North American ice melted about 12,000 years ago: pine siskin would have followed the ancestral North to South migrations observed today and covered all North America. It might occupy American niches of Eurasian siskin which could not reach America from Asia during the last 2 million years because of an extant thick ice shield. Neither could it later because of species competition by ecologic niche with its descent pine siskin [12Arnaiz-Villena A, Ruiz-del-Valle V, Reguera R, Gomez-Prieto P, Serrano-Vela JI. What might have been the ancestor of New World siskins? Open Ornithol J 2008; 1: 46-7.
[http://dx.doi.org/10.2174/1874453200801010046] ].
4.2. Number of Alleles
A detailed genetic map of the MHC in songbirds has not been obtained, so nothing can be said about the precise number of genes that composes it and its proximity on the chromosome. In spite of this, some gene numbers have been postulated [43Westerdhal H. Passerine MHC: Genetic variation and disease resistance in the wild. J Ornithol 2007; 148: 469-77.
[http://dx.doi.org/10.1007/s10336-007-0230-5] ]. In this work we have not analyzed such a characteristic and we have found no more than two different MHC alleles per single individual of the studied species of these particular genes. Therefore, our findings fit with detection of one paternal and one maternal gene belonging to a single locus.
In species such as Coturnix japonica at least four classic class I genes have been found with a high variety of alleles [44Shiina T, Ando A, Imanishi T, et al. Isolation and characterization of cDNA clones for Japanese quail (Coturnix japonica) major histocompatibility complex (MhcCoja) class I molecules. Immunogenetics 1995; 42(3): 213-6.
[http://dx.doi.org/10.1007/BF00191227] [PMID: 7642233] -46Shiina T, Shimizu S, Hosomichi K, et al. Comparative genomic analysis of two avian (quail and chicken) MHC regions. J Immunol 2004; 172(11): 6751-63.
[http://dx.doi.org/10.4049/jimmunol.172.11.6751] [PMID: 15153492] ]; this also is the case of goose [47Xia C, Hu T, Yang T, et al. cDNA cloning, genomic structure and expression analysis of the goose (Anser cygnoides) MHC class I gene. Vet Immunol Immunopathol 2005; 107(3-4): 291-302.
[http://dx.doi.org/10.1016/j.vetimm.2005.05.005] [PMID: 16005079] ]. Duck has five classic class I genes although only two of them seem to actively work [48Moon DA, Veniamin SM, Parks-Dely JA, Magor KE. The MHC of the duck (Anas platyrhynchos) contains five differentially expressed class I genes. J Immunol 2005; 175(10): 6702-12.
[http://dx.doi.org/10.4049/jimmunol.175.10.6702] [PMID: 16272326] ]. In songbirds, class I sequences have been studied in very few species. In the great reed warbler [49Westerdahl H, Wittzell H, von Schantz T. Polymorphism and transcription of Mhc class I genes in a passerine bird, the great reed warbler. Immunogenetics 1999; 49(3): 158-70.
[http://dx.doi.org/10.1007/s002510050477] [PMID: 9914330] , 50Wittzell H, Bernot A, Auffray C, Zoorob R. Concerted evolution of two Mhc class II B loci in pheasants and domestic chickens. Mol Biol Evol 1999; 16(4): 479-90.
[http://dx.doi.org/10.1093/oxfordjournals.molbev.a026130] [PMID: 10331274] ] a high genetic MHC variability has been described compared with chicken (Gallus gallus) while in South American siskins [51Ferre S. Evolución de los genes de histocompatibilidad de clase I en la radiación de jilgueros (lúganos) sudamericanos [Doctoral Thesis] 2001.] and in canaries [52Lowy E. Evolución y Sistema Principal de Histocompatibilidad en canarios (Género Serinus) [Doctoral Thesis] 2003., 53Arnaiz-Villena A, Lowy E, Ruiz-del-Valle V, et al. Evolution of the major histocompatibility complex class I genes in Serinus canaria from the Canary Islands is different from that of Asian and African continental Serinus species. J Ornithol 2007; 148: 479-84.
[http://dx.doi.org/10.1007/s10336-007-0146-0] ] only one gene with a low variability has been found.
4.3. MHC Transpecies Evolution in Birds
A transpecific gene existance occurs in several mammals MHC, like apes [25Suárez B, Morales P, Castro MJ, et al. Mhc-E polymorphism in Pongidae primates: The same allele is found in two different species. Tissue Antigens 1997; 50(6): 695-8.
[http://dx.doi.org/10.1111/j.1399-0039.1997.tb02938.x] [PMID: 9458133] , 51Ferre S. Evolución de los genes de histocompatibilidad de clase I en la radiación de jilgueros (lúganos) sudamericanos [Doctoral Thesis] 2001.]. This phenomenon usually occurs when speciation happens quickly, while gene differentiation has not yet taken place. It could also mean that the MHC naturally adapts to habitat of species and select alleles to combat characteristic antigens / pathogens thriving in the area, and does not need to generate an unlimited polymorphism as in the case of “artificial” MHC, where there are a high number of alleles and numerous immunological disorders that appear to be associated with the HLA system, such as autoimmune processes [51Ferre S. Evolución de los genes de histocompatibilidad de clase I en la radiación de jilgueros (lúganos) sudamericanos [Doctoral Thesis] 2001., 52Lowy E. Evolución y Sistema Principal de Histocompatibilidad en canarios (Género Serinus) [Doctoral Thesis] 2003.]. Human, laboratory mouse and chicken are considered “artificial” vertebrate models because all have originated through a bottleneck and subsequent relatively high inbreeding, which enhances crossover at meiosis and thus to excessive MHC diversity [22Klein J, Sato A, Nagl S. O’hUigin C. Molecular trans-species polymorphism. Annu Rev Ecol Syst 1998; 29: 1-21.
[http://dx.doi.org/10.1146/annurev.ecolsys.29.1.1] , 51Ferre S. Evolución de los genes de histocompatibilidad de clase I en la radiación de jilgueros (lúganos) sudamericanos [Doctoral Thesis] 2001., 52Lowy E. Evolución y Sistema Principal de Histocompatibilidad en canarios (Género Serinus) [Doctoral Thesis] 2003.].
Phylogenetic analysis of MHC sequences from the species studied in the present work allowed to visualizing more clearly the phenomenon of trans-specificity since, two matches between MHC-I alleles from different South American siskin species were found at DNA level. It was also found that the Eurasian siskin shares a MHC protein allele with the pine siskin (North-Central America range) and another protein allele with three South American siskin species. Eight South American siskins species, including parental C. notata, also share another MHC protein among themselves (Fig. 3 ). All MHC genes transmit their alleles to the descendant species, but the most common fact is that the allelic identity between the ancestral species and their descendants get lost due to balancing selection diversification. The antiquity of studied MHC alleles goes back no longer than four - five millions years, when South American siskins and North American goldfinches species were separated (Figs. 1
). All MHC genes transmit their alleles to the descendant species, but the most common fact is that the allelic identity between the ancestral species and their descendants get lost due to balancing selection diversification. The antiquity of studied MHC alleles goes back no longer than four - five millions years, when South American siskins and North American goldfinches species were separated (Figs. 1 and 2
and 2 ) [11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
) [11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
[http://dx.doi.org/10.2174/1874453201205010073] ].
A closest relationship between the Eurasian siskin and the Pine siskin had been found [8Arnaiz-Villena A, Alvarez-Tejado M, Ruíz-del-Valle V, et al. Phylogeny and rapid northern and southern hemisphere speciation of goldfinches during the Miocene and Pliocene epochs. Cell Mol Life Sci 1998; 54(9): 1031-41.
[http://dx.doi.org/10.1007/s000180050230] [PMID: 9791543] , 10Arnaiz-Villena A, Ruiz-del-Valle V, Moscoso J, Serrano-Vela JI, Zamora J. mtDNA phylogeny of North American Carduelis pinus group. Ardeola 2007; 54: 1-14., 11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
[http://dx.doi.org/10.2174/1874453201205010073] ]. This is further supported by the fact of sharing one MHC protein allele. Eurasian siskin (o extant relative) could have been a species widely spread around Europe, Asia and America that could be have led to both North American goldfinches and South American siskins radiations. Otherwise, it could have only originated North American goldfinches radiations and these could have originated the South American siskin radiation [11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
[http://dx.doi.org/10.2174/1874453201205010073] , 12Arnaiz-Villena A, Ruiz-del-Valle V, Reguera R, Gomez-Prieto P, Serrano-Vela JI. What might have been the ancestor of New World siskins? Open Ornithol J 2008; 1: 46-7.
[http://dx.doi.org/10.2174/1874453200801010046] ].
In conclusion, our data on mitochondrial cytochrome-b combined with the first evidence of trans-species MHC evolution so far described in birds, suggests that the Eurasian siskin is the extant ancestor of all North and South American Carduelis species [11Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81.
[http://dx.doi.org/10.2174/1874453201205010073] , 12Arnaiz-Villena A, Ruiz-del-Valle V, Reguera R, Gomez-Prieto P, Serrano-Vela JI. What might have been the ancestor of New World siskins? Open Ornithol J 2008; 1: 46-7.
[http://dx.doi.org/10.2174/1874453200801010046] ].
4.4. MHC Large Intron Size in Passerines
This set of wild bird species studied (Table 1 [26Arnaiz-Villena A, Ruiz-del-Valle V, Reche P, et al. Songbirds conserved sites and intron size of MHC Class I molecules reveal a unique evolution in vertebrates. Open Ornithol J 2010; 3: 156-65.
[http://dx.doi.org/10.2174/1874453201003010156] ],) has given the first direct evidence that one of the main characters of “Minimal Essential Bird MHC” postulated for birds [54Kaufman J, Milne S, Göbel TW, et al. The chicken B locus is a minimal essential major histocompatibility complex. Nature 1999; 401(6756): 923-5.
[http://dx.doi.org/10.1038/44856] [PMID: 10553909] ] is in fact not universal for birds.
MHC class I introns from presently and previously [26Arnaiz-Villena A, Ruiz-del-Valle V, Reche P, et al. Songbirds conserved sites and intron size of MHC Class I molecules reveal a unique evolution in vertebrates. Open Ornithol J 2010; 3: 156-65.
[http://dx.doi.org/10.2174/1874453201003010156] ] studied songbirds are longer than humans. Chicken introns are the shortest ones [26Arnaiz-Villena A, Ruiz-del-Valle V, Reche P, et al. Songbirds conserved sites and intron size of MHC Class I molecules reveal a unique evolution in vertebrates. Open Ornithol J 2010; 3: 156-65.
[http://dx.doi.org/10.2174/1874453201003010156] ]. In addition, MHC class I genes introns 2 were homologous on 38.3% to human class I MHC and 35% to Gallus gallus one. MHC class I intron 2 had an average of 304 bp in songbirds, 238 bp in human and 288 bp in Gallus gallus [26Arnaiz-Villena A, Ruiz-del-Valle V, Reche P, et al. Songbirds conserved sites and intron size of MHC Class I molecules reveal a unique evolution in vertebrates. Open Ornithol J 2010; 3: 156-65.
[http://dx.doi.org/10.2174/1874453201003010156] ]. This is also a direct evidence that the main postulate of “Minimal Essential MHC” for birds only applies to Gallus gallus, and not to Passerines [54Kaufman J, Milne S, Göbel TW, et al. The chicken B locus is a minimal essential major histocompatibility complex. Nature 1999; 401(6756): 923-5.
[http://dx.doi.org/10.1038/44856] [PMID: 10553909] ]. Additional information may be found in references [55Arnaiz-Villena A, Ruiz-del-Valle V, Gomez-Prieto P, et al. Carduelini New Sistematics: Crimson-winged Finch (Rhodopechys sanguineus) is Included in “Arid-Zone” Carduelini Finches by Mitochondrial DNA Phylogeny. Open Ornithol J 2014; 7: 55-62.
[http://dx.doi.org/10.2174/1874453201407010055] , 56Karlsson M, Westerdahl H. Characteristics of MHC class I genes in house sparrows Passer domesticus as revealed by long cDNA transcripts and amplicon sequencing. J Mol Evol 2013; 77(1-2): 8-21.
[http://dx.doi.org/10.1007/s00239-013-9575-y] [PMID: 23877344] ].The fact that songbirds other than Carduelinae family (i.e.: Acrocephalus arundinaceus and Taeniopygia guttata, zebra finch [26Arnaiz-Villena A, Ruiz-del-Valle V, Reche P, et al. Songbirds conserved sites and intron size of MHC Class I molecules reveal a unique evolution in vertebrates. Open Ornithol J 2010; 3: 156-65.
[http://dx.doi.org/10.2174/1874453201003010156] ]) are also different at residues 10 and 96 at MHC class I proteins than all other vertebrates (including Gallus gallus) may indicate that our observations of on large intron size and different conserved 10 and 96 residues on MHC proteins could be extended to songbird family (i.e.: about half of the about 10,000 avian extant species [3Sibley CG, Monroe BL. Distribution and Taxonomy of Birds of the World 1990.]). Passerine evolutionary pathway may be altogether different from that of other birds [26Arnaiz-Villena A, Ruiz-del-Valle V, Reche P, et al. Songbirds conserved sites and intron size of MHC Class I molecules reveal a unique evolution in vertebrates. Open Ornithol J 2010; 3: 156-65.
[http://dx.doi.org/10.2174/1874453201003010156] ].
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This project was approved by University Complutense Ethics Committee.
HUMAN AND ANIMAL RIGHTS
The reported experiments in accordance with the standards set forth in the 8th Edition of Guide for the Care and Use of Laboratory Animal (http://grants.nih.gov/grants/olaw/Guide-for-the-care-and-use-of-laboratory-animals.pdf) published by the National Academy of Sciences, The National Academies Press, Washington DC, United States of America..
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
This work was supported in part by Grants from the Spanish Ministry of Health and Economy and European FEDER funds (PI14/01067).
REFERENCES
| [1] | Arnaiz-Villena A, Guillén J, Ruiz-del-Valle V, et al. Phylogeography of crossbills, bullfinches, grosbeaks, and rosefinches. Cell Mol Life Sci 2001; 58(8): 1159-66. [http://dx.doi.org/10.1007/PL00000930] [PMID: 11529508] |
| [2] | Armani GC. Guide des Passereaux Granivores 1983. |
| [3] | Sibley CG, Monroe BL. Distribution and Taxonomy of Birds of the World 1990. |
| [4] | Clement P, Harris A, Davis J. Finches and Sparrows 1999. |
| [5] | Arnaiz-Villena A, Alvarez-Tejado M, Ruiz-del-Valle V, et al. Rapid radiation of canaries (Genus Serinus). Mol Biol Evol 1999; 16: 2-11. [http://dx.doi.org/10.1093/oxfordjournals.molbev.a026034] |
| [6] | Allende LM, Rubio I, Ruíz-Del-Valle V, et al. The Old World sparrows (genus Passer) phylogeography and their relative abundance of nuclear mtDNA pseudogenes. J Mol Evol 2001; 53(2): 144-54. [http://dx.doi.org/10.1007/s002390010202] [PMID: 11479685] |
| [7] | Arnaiz-Villena A, Gomez-Prieto P, Ruiz-del-Valle V. Phylogeography of finches and sparrows 2009; 1-54. |
| [8] | Arnaiz-Villena A, Alvarez-Tejado M, Ruíz-del-Valle V, et al. Phylogeny and rapid northern and southern hemisphere speciation of goldfinches during the Miocene and Pliocene epochs. Cell Mol Life Sci 1998; 54(9): 1031-41. [http://dx.doi.org/10.1007/s000180050230] [PMID: 9791543] |
| [9] | Helm-Bychowski KM, Wilson AC. Rates of nuclear DNA evolution in pheasant-like birds: evidence from restriction maps. Proc Natl Acad Sci USA 1986; 83(3): 688-92. [http://dx.doi.org/10.1073/pnas.83.3.688] [PMID: 3003745] |
| [10] | Arnaiz-Villena A, Ruiz-del-Valle V, Moscoso J, Serrano-Vela JI, Zamora J. mtDNA phylogeny of North American Carduelis pinus group. Ardeola 2007; 54: 1-14. |
| [11] | Arnaiz-Villena A, Areces C, Rey D, et al. Three different North American Siskin/Goldfinch Evolutionary Radiations (Genus Carduelis): Pine Siskin Green Morphs and European siskins in America. Open Ornithol J 2012; 5: 73-81. [http://dx.doi.org/10.2174/1874453201205010073] |
| [12] | Arnaiz-Villena A, Ruiz-del-Valle V, Reguera R, Gomez-Prieto P, Serrano-Vela JI. What might have been the ancestor of New World siskins? Open Ornithol J 2008; 1: 46-7. [http://dx.doi.org/10.2174/1874453200801010046] |
| [13] | Klein J. Natural history of the Major Histocompatibility Complex 1986. |
| [14] | Murphy K, Travers P, Walport M. Janeway’s immunobiology 2008. |
| [15] | Gomez-Prieto P, Parga-Lozano C, Rey D, Moreno E, Arnaiz-Villena A. HLA-G, -F and -E: polymorphism, function and evolution 2010; 159-74. |
| [16] | Donadi EA, Castelli EC, Arnaiz-Villena A, et al. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci 2011; 68(3): 369-95. [http://dx.doi.org/10.1007/s00018-010-0580-7] [PMID: 21107637] |
| [17] | Graur D, Li WH. Fundamentals of molecular evolution 2nd ed. 1999. |
| [18] | Lewontin RC, Hubby JL. A molecular approach to the study of genic heterozygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics 1966; 54(2): 595-609. [PMID: 5968643] |
| [19] | Doherty PC, Zinkernagel RM. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature 1975; 256(5512): 50-2. [http://dx.doi.org/10.1038/256050a0] [PMID: 1079575] |
| [20] | Black FL, Salzano FM. Evidence for heterosis in the HLA system. Am J Hum Genet 1981; 33(6): 894-9. [PMID: 7325154] |
| [21] | Bodmer WF, Bodmer JG. Evolution and function of the HLA system. Br Med Bull 1978; 34(3): 309-16. [http://dx.doi.org/10.1093/oxfordjournals.bmb.a071518] [PMID: 363227] |
| [22] | Klein J, Sato A, Nagl S. O’hUigin C. Molecular trans-species polymorphism. Annu Rev Ecol Syst 1998; 29: 1-21. [http://dx.doi.org/10.1146/annurev.ecolsys.29.1.1] |
| [23] | Arnaiz-Villena A, Enriquez-de-Salamanca M, Palacio-Grüber J, et al. Characterisation and functional implications of the two new HLA-G alleles found in Amerindian and Caribbean populations. Hum Immunol 2016; 77(9): 812-6. [http://dx.doi.org/10.1016/j.humimm.2016.01.006] [PMID: 26796363] |
| [24] | Arnaiz-Villena A, Morales P, Gomez-Casado E, et al. Evolution of MHC-G in primates: A different kind of molecule for each group of species. J Reprod Immunol 1999; 43(2): 111-25. [http://dx.doi.org/10.1016/S0165-0378(99)00026-1] [PMID: 10479048] |
| [25] | Suárez B, Morales P, Castro MJ, et al. Mhc-E polymorphism in Pongidae primates: The same allele is found in two different species. Tissue Antigens 1997; 50(6): 695-8. [http://dx.doi.org/10.1111/j.1399-0039.1997.tb02938.x] [PMID: 9458133] |
| [26] | Arnaiz-Villena A, Ruiz-del-Valle V, Reche P, et al. Songbirds conserved sites and intron size of MHC Class I molecules reveal a unique evolution in vertebrates. Open Ornithol J 2010; 3: 156-65. [http://dx.doi.org/10.2174/1874453201003010156] |
| [27] | Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol 1987; 155: 335-50. [http://dx.doi.org/10.1016/0076-6879(87)55023-6] [PMID: 3431465] |
| [28] | Saiki RK, Scharf S, Faloona F, et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 1985; 230(4732): 1350-4. [http://dx.doi.org/10.1126/science.2999980] [PMID: 2999980] |
| [29] | Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 1977; 74(12): 5463-7. [http://dx.doi.org/10.1073/pnas.74.12.5463] [PMID: 271968] |
| [30] | Zamora J, Lowy E, Ruiz-del-Valle V, et al. Rhodopechys obsoleta (desert finch): A pale ancestor of greenfinches (Carduelis spp.) according to molecular phylogeny. J Ornithol 2006; 147: 448-56. [http://dx.doi.org/10.1007/s10336-005-0036-2] |
| [31] | Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28(10): 2731-9. [http://dx.doi.org/10.1093/molbev/msr121] [PMID: 21546353] |
| [32] | Felsenstein J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J Mol Evol 1981; 17(6): 368-76. [http://dx.doi.org/10.1007/BF01734359] [PMID: 7288891] |
| [33] | Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (* and other methods) version 4 2002. |
| [34] | Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001; 17(8): 754-5. [http://dx.doi.org/10.1093/bioinformatics/17.8.754] [PMID: 11524383] |
| [35] | Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003; 19(12): 1572-4. [http://dx.doi.org/10.1093/bioinformatics/btg180] [PMID: 12912839] |
| [36] | Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics 1998; 14(9): 817-8. [http://dx.doi.org/10.1093/bioinformatics/14.9.817] [PMID: 9918953] |
| [37] | Thorne JL, Kishino H, Painter IS. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol 1998; 15(12): 1647-57. [http://dx.doi.org/10.1093/oxfordjournals.molbev.a025892] [PMID: 9866200] |
| [38] | McLaren IA, Morlan J, Smith P, Gosselin M, Bailey SF. Eurasian siskins in North America - distinguishing females from green-morp pine siskins. American Birds 1989; 43: 1268-74. |
| [39] | Klein J, Bontrop RE, Dawkins RL, et al. Nomenclature for the major histocompatibility complexes of different species: A proposal. Immunogenetics 1990; 31(4): 217-9. [http://dx.doi.org/10.1007/BF00204890] [PMID: 2329006] |
| [40] | Bonneaud C, Sorci G, Morin V, et al. Diversity of Mhc class I and IIB genes in house sparrows (Passer domesticus). Immunogenetics 2004; 55(12): 855-65. [http://dx.doi.org/10.1007/s00251-004-0648-3] [PMID: 14963619] |
| [41] | Lomolino MV, Riddle BR, Brown JH. Biogeography 3rd ed. 2006. |
| [42] | Pregill GK, Olson SL. Zoogeography of west indian vertebrates in relation to pleistocene climatic cycles. Annu Rev Ecol Syst 1981; 12: 75-98. [http://dx.doi.org/10.1146/annurev.es.12.110181.000451] |
| [43] | Westerdhal H. Passerine MHC: Genetic variation and disease resistance in the wild. J Ornithol 2007; 148: 469-77. [http://dx.doi.org/10.1007/s10336-007-0230-5] |
| [44] | Shiina T, Ando A, Imanishi T, et al. Isolation and characterization of cDNA clones for Japanese quail (Coturnix japonica) major histocompatibility complex (MhcCoja) class I molecules. Immunogenetics 1995; 42(3): 213-6. [http://dx.doi.org/10.1007/BF00191227] [PMID: 7642233] |
| [45] | Shiina T, Shimizu C, Oka A, et al. Gene organization of the quail major histocompatibility complex (MhcCoja) class I gene region. Immunogenetics 1999; 49(5): 384-94. [http://dx.doi.org/10.1007/s002510050511] [PMID: 10199914] |
| [46] | Shiina T, Shimizu S, Hosomichi K, et al. Comparative genomic analysis of two avian (quail and chicken) MHC regions. J Immunol 2004; 172(11): 6751-63. [http://dx.doi.org/10.4049/jimmunol.172.11.6751] [PMID: 15153492] |
| [47] | Xia C, Hu T, Yang T, et al. cDNA cloning, genomic structure and expression analysis of the goose (Anser cygnoides) MHC class I gene. Vet Immunol Immunopathol 2005; 107(3-4): 291-302. [http://dx.doi.org/10.1016/j.vetimm.2005.05.005] [PMID: 16005079] |
| [48] | Moon DA, Veniamin SM, Parks-Dely JA, Magor KE. The MHC of the duck (Anas platyrhynchos) contains five differentially expressed class I genes. J Immunol 2005; 175(10): 6702-12. [http://dx.doi.org/10.4049/jimmunol.175.10.6702] [PMID: 16272326] |
| [49] | Westerdahl H, Wittzell H, von Schantz T. Polymorphism and transcription of Mhc class I genes in a passerine bird, the great reed warbler. Immunogenetics 1999; 49(3): 158-70. [http://dx.doi.org/10.1007/s002510050477] [PMID: 9914330] |
| [50] | Wittzell H, Bernot A, Auffray C, Zoorob R. Concerted evolution of two Mhc class II B loci in pheasants and domestic chickens. Mol Biol Evol 1999; 16(4): 479-90. [http://dx.doi.org/10.1093/oxfordjournals.molbev.a026130] [PMID: 10331274] |
| [51] | Ferre S. Evolución de los genes de histocompatibilidad de clase I en la radiación de jilgueros (lúganos) sudamericanos [Doctoral Thesis] 2001. |
| [52] | Lowy E. Evolución y Sistema Principal de Histocompatibilidad en canarios (Género Serinus) [Doctoral Thesis] 2003. |
| [53] | Arnaiz-Villena A, Lowy E, Ruiz-del-Valle V, et al. Evolution of the major histocompatibility complex class I genes in Serinus canaria from the Canary Islands is different from that of Asian and African continental Serinus species. J Ornithol 2007; 148: 479-84. [http://dx.doi.org/10.1007/s10336-007-0146-0] |
| [54] | Kaufman J, Milne S, Göbel TW, et al. The chicken B locus is a minimal essential major histocompatibility complex. Nature 1999; 401(6756): 923-5. [http://dx.doi.org/10.1038/44856] [PMID: 10553909] |
| [55] | Arnaiz-Villena A, Ruiz-del-Valle V, Gomez-Prieto P, et al. Carduelini New Sistematics: Crimson-winged Finch (Rhodopechys sanguineus) is Included in “Arid-Zone” Carduelini Finches by Mitochondrial DNA Phylogeny. Open Ornithol J 2014; 7: 55-62. [http://dx.doi.org/10.2174/1874453201407010055] |
| [56] | Karlsson M, Westerdahl H. Characteristics of MHC class I genes in house sparrows Passer domesticus as revealed by long cDNA transcripts and amplicon sequencing. J Mol Evol 2013; 77(1-2): 8-21. [http://dx.doi.org/10.1007/s00239-013-9575-y] [PMID: 23877344] |