- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Molecular Characterization of BK Polyomavirus’ Large T Antigen Gene Sequences Detected in Prostate Cancer Tissues of Sudanese Patients
Babbiker M. T. Gorish1, *, Mohammed E. H. Ournasseir1, Iman M. Shammat1, 2
Abstract
Background:
BK virus, which is associated with Prostate Cancer (PCa), have a global seroprevalence in humans. Based on the sequences of VP1 and the Large Antigen (LTAg) genes, there are four subtypes of BKV. Each subtype has its own subgroups.
Objective:
The aim of this study was to identify the BKV subtype that circulates among Sudanese patients with PCa.
Materials and Methods:
A total of 8 samples from our previous work on BKV were studied in this investigation. The LTAg gene was partially amplified (176nt) by a homemade PCR. All the amplicons were purified and subjected to sequencing. Bioedit version 7.0 and Mega X version 6.0 were used to analyze the sequence and compare the results with the BKV sequences and build a phylogenetic tree.
Results:
All the BKV LTAg gene sequences derived from Sudanese patients were classified with Subtype-1 BKV strains from Iran and Japan. Translated protein alignment showed that some isolates had identical amino acids with Iranian and Japanese strains, whereas others had a silent mutation. Interestingly, a point mutation was identified in the sequences of isolate 5 and 8 where adenine nucleotide (A) was replaced with Cytosine (C) at position 276, resulting in amino acid substitution.
Conclusion:
It was concluded that all the BKV isolates which circulated among Sudanese prostate tumor patients belonged to subtype 1. These findings only highlighted the need for the molecular detection and subtyping of BKV strains in Sudanese patients in order to better demonstrate the relationship between BKV infection and PCa.
Article Information
Identifiers and Pagination:
Year: 2019Volume: 13
First Page: 29
Last Page: 37
Publisher Id: TOVJ-13-29
DOI: 10.2174/1874357901913010029
Article History:
Received Date: 11/07/2019Revision Received Date: 19/10/2019
Acceptance Date: 31/10/2019
Electronic publication date: 20/12/2019
Collection year: 2019
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: (https://creativecommons.org/licenses/by/4.0/legalcode). This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
* Address correspondence to this author at the Department of Microbiology and Immunology, Faculty of Medical Laboratory Science, Omdurman Islamic University, Omdurman, Sudan; E-mail: qorish456@gmail.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 11-07-2019 |
Original Manuscript | Molecular Characterization of BK Polyomavirus’ Large T Antigen Gene Sequences Detected in Prostate Cancer Tissues of Sudanese Patients | |
1. INTRODUCTION
The prostate gland is one of the most essential male accessory glands. This gland is susceptible to various pathological conditions, among which both malignant and benign conditions are the most common [1Verhamme KM, Dieleman JP, Bleumink GS, et al. Triumph Pan European Expert Panel. Incidence and prevalence of lower urinary tract symptoms suggestive of benign prostatic hyperplasia in primary care--the Triumph project. Eur Urol 2002; 42(4): 323-8.
[http://dx.doi.org/10.1016/S0302-2838(02)00354-8] [PMID: 12361895] ]. Benign Prostatic Hyperplasia (BPH), which is not a cancer, is common among older men and occurs when the prostate gland is enlarged. When the gland becomes larger it ‘squeezes’ the urethra, thus causing several complications, such as difficulty urinating and frequent urination during the day [2Grossman DC, Curry SJ, Owens DK, et al. US Preventive Services Task Force. Screening for Prostate Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2018; 319(18): 1901-13.
[http://dx.doi.org/10.1001/jama.2018.3710] [PMID: 29801017] ]. PCa is a biologically heterogeneous tumor and is one of the leading causes of cancer death in men [3Rodríguez H, Levican J, Muñoz JP, et al. Viral infections in prostate carcinomas in Chilean patients. Infect Agent Cancer 2015; 10: 27.
[http://dx.doi.org/10.1186/s13027-015-0024-y] [PMID: 26330890] ]. 241,740 new cases and 28,170 deaths were estimated in 2012 [4Delbue S, Matei DV, Carloni C, et al. Evidence supporting the association of polyomavirus BK genome with prostate cancer. Med Microbiol Immunol (Berl) 2013; 202(6): 425-30.
[http://dx.doi.org/10.1007/s00430-013-0304-3] [PMID: 23821367] ]. It is the second leading cause of cancer-related death in men in the United States [5Elimam ME, Abdelraof S. Incidence of carcinoma of the prostate in patients with normal prostatic specific antigen following prostatectomy. Glob J Medi Res 2013; 13: 26-35.]. The underlying causes of PCa are not fully understood, but it is likely to occur due to a combination of factors, such as aging, family history and dietary factors in addition to infectious agents [4Delbue S, Matei DV, Carloni C, et al. Evidence supporting the association of polyomavirus BK genome with prostate cancer. Med Microbiol Immunol (Berl) 2013; 202(6): 425-30.
[http://dx.doi.org/10.1007/s00430-013-0304-3] [PMID: 23821367] -6Provenzano M, Keller P. The potential therapeutic usefulness of targeting BK polyoma virus in prostate cancer. Chemo Open Access 2015; 5: 1.]. Pooled data from population studies on PCa risk have reported an 80% increase in the risk of PCA in men with a history of prostatitis, although the detection of potential bias plays a role in it. Proliferative Inflammatory Atrophy (PIA) is common in normal and cancerous prostate and is a regenerative lesion after injury or trauma. It has been suggested that PIA is a precursor lesion of PCa and several genetic mutations have been identified in PIA, including glutathione S-transferase pi 1 (GSTP1) hypermethylation, p53 mutations, and alterations on chromosome 8 [7Martinez-Fierro ML, Leach RJ, Gomez-Guerra LS, et al. Identification of viral infections in the prostate and evaluation of their association with cancer. BMC Cancer 2010; 10: 326.
[http://dx.doi.org/10.1186/1471-2407-10-326] [PMID: 20576103] ].
Several pieces of evidence support an infectious pathway in PCa development. In particular, Sexually Transmitted Infections (STIs) have been implicated in PCa etiology in many studies. A meta-analysis of 29 case-control studies found an increased relative risk of PCa in men with a history of any STI with an odds ratio of 1.5 and 95% Confidence Interval (CI), (1.3-1.7). Several sexually transmitted organisms have been detected in the prostate, including Neisseria gonorrhoeae, Chlamydia trachomatis, Trichomonas vaginalis, Treponema pallidum, human papillomavirus, herpes simplex virus, and human herpesvirus type 8. Serum antibodies against herpes simplex virus and Trichomonas vaginalis in men have been associated with PCa risk, whereas the presence of the DNA of human papillomavirus in prostate tissues has also been associated with the risk of PCa [8Wright JL, Lin DW, Stanford JL. Circumcision and the risk of prostate cancer. Cancer 2012; 118(18): 4437-43.
[http://dx.doi.org/10.1002/cncr.26653] [PMID: 22411189] ]. Infectious agents may lead to cancer by several potential mechanisms. Although, direct cellular transformation can occur by viruses, several changes in the tissue microenvironment can occur if a chronic inflammatory state is reached after the infection. These include damage due to reactive oxygen species along with cytokine-induced angiogenesis and cellular proliferation, all of which may be involved in carcinogenesis [9De Marzo AM, Platz EA, Sutcliffe S, et al. Inflammation in prostate carcinogenesis. Nat Rev Cancer 2007; 7(4): 256-69.
[http://dx.doi.org/10.1038/nrc2090] [PMID: 17384581] ].
The physiopathology of BKV-induced inflammation and the role of a cofactor in prostate and other cancers can be summarized by the fact that BKV T antigens, such as JCV, display multiple functions which alter the normal physiological metabolism of cells, ultimately leading to immortalization and neoplastic transformation. The main property of Tag in relation to the transformation and oncogenicity is its ability to bind and block the functions of tumor suppressor proteins p53 and pRB family (p105 RB1, p107 and p130 RB2). BKV Tag also induces numerical and structural chromosomal alterations in human embryonic fibroblasts, characterized by gaps, breaks, dicentric and ring chromosomes, deletions, duplications and translocations. Chromosome damage in human cells transfected with BKV early region was evident before the appearance of immortalization and the morphologically transformed phenotype [10Das D, Wojno K, Imperiale MJ. BK virus as a cofactor in the etiology of prostate cancer in its early stages. J Virol 2008; 82(6): 2705-14.
[http://dx.doi.org/10.1128/JVI.02461-07] [PMID: 18160432] -13Zambrano A, Kalantari M, Simoneau A, Jensen JL, Villarreal LP. Detection of human polyomaviruses and papillomaviruses in prostatic tissue reveals the prostate as a habitat for multiple viral infections. Prostate 2002; 53(4): 263-76.
[http://dx.doi.org/10.1002/pros.10157] [PMID: 12430138] ].
The human BK polyomavirus is a member of the polyomavirus family. The virus has an icosahedral shaped capsid which is unenveloped, measuring about 40-50 nm. The capsid encloses a circular double-stranded DNA genome of approximately 5100 nucleotides coated by host-cell histones [14Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet 1971; 1(7712): 1253-7.
[http://dx.doi.org/10.1016/S0140-6736(71)91776-4] [PMID: 4104714] ]. The virus was first isolated from the urine of a renal transplant patient in 1971 [14Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet 1971; 1(7712): 1253-7.
[http://dx.doi.org/10.1016/S0140-6736(71)91776-4] [PMID: 4104714] ] and infects almost 90% of the human population worldwide. It rarely causes diseases in immune-competent individuals and resides in a subclinical persistent state in the urinary tract of healthy individuals and reactivates in immunosuppressed transplant patients, in whom it is associated with certain diseases, such as hemorrhagic cystitis and polyomavirus nephropathy [15Tognon M, Corallini A, Martini F, Negrini M, Barbanti-Brodano G. Oncogenic transformation by BK virus and association with human tumors. Oncogene 2003; 22(33): 5192-200.
[http://dx.doi.org/10.1038/sj.onc.1206550] [PMID: 12910256] ]. Urinary shedding has been reported to occur asymptomatically and intermittently in healthy individuals [16Dalianis T, Hirsch HH. Human polyomaviruses in disease and cancer. Virology 2013; 437(2): 63-72.
[http://dx.doi.org/10.1016/j.virol.2012.12.015] [PMID: 23357733] ].
The genome of BKV is divided into two transcriptional regions called early and late regions in addition to the third non-transcriptional region called the regulatory region. The genome of a typical BK polyomavirus codes for between 5 and 9 proteins, two from the early region and the rest from the late region. The early region is transcripted by the host cell's RNA polymerase II and codes for two proteins, the small and large tumor antigens, produced by alternative splicing [17Ahuja D, Sáenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 2005; 24(52): 7729-45.
[http://dx.doi.org/10.1038/sj.onc.1209046] [PMID: 16299533] ]. The Large Tumor Antigen (LTAg) is expressed early in the infectious cycle and is essential for virus proliferation. It is about 600-800 amino acids in length containing four well-conserved protein domains as well as several intrinsically disordered regions. The antigen has two primary functions, both related to virus replication. Its first function is to unwind the viral DNA and prepare it for replication, while the second is to destroy the cell cycle by interaction with the host cell protein. In addition, the LTAg promotes cellular transformation by interfering with the tumor suppressor functions of Rb, p107, p130, and p53 (Fig. 1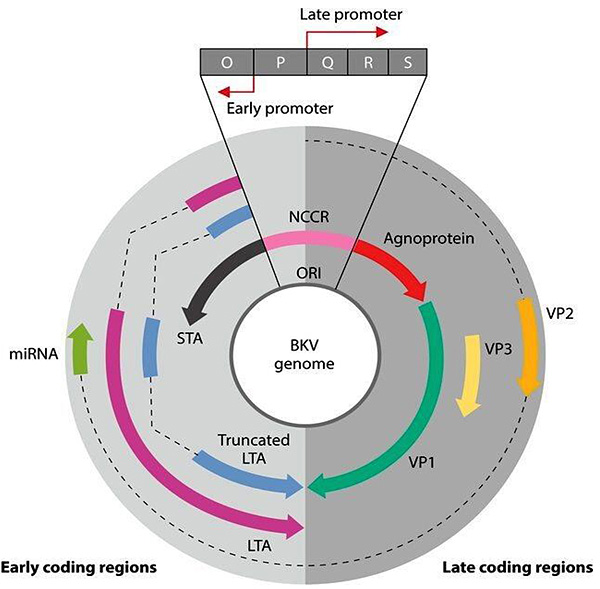 ) [17Ahuja D, Sáenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 2005; 24(52): 7729-45.
) [17Ahuja D, Sáenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 2005; 24(52): 7729-45.
[http://dx.doi.org/10.1038/sj.onc.1209046] [PMID: 16299533] -19Maurizio P, Etienne XK. The potential therapeutic usefulness of targeting BK Polyomavirusin prostate cancer. Chemo Open Access 2015; 5: 1-2.]. Another protein that is expressed early in the BKV infectious cycle is the small Tumor Antigen (TAg). Unlike the LTAg, it is not essential for viral replication, instead it induces tumorogenesis and promotes anchorage-independent growth of transformed cells by interaction with the cell protein phosphatase 2A (Fig. 1 ) [18Yu J, Boyapati A, Rundell K. Critical role for SV40 small-t antigen in human cell transformation. Virology 2001; 290(2): 192-8.
) [18Yu J, Boyapati A, Rundell K. Critical role for SV40 small-t antigen in human cell transformation. Virology 2001; 290(2): 192-8.
[http://dx.doi.org/10.1006/viro.2001.1204] [PMID: 11883184] , 19Maurizio P, Etienne XK. The potential therapeutic usefulness of targeting BK Polyomavirusin prostate cancer. Chemo Open Access 2015; 5: 1-2.]
The first genotyping schema for BKV described by Jin et al. in 1993 was based on a very short segment of the VP1 gene (nucleotides 1744 to 1812). However, phylogenetic trees based on LTA sequences were introduced and validated recently [20Luo C, Bueno M, Kant J, Martinson J, Randhawa P. Genotyping schemes for polyomavirus BK, using gene-specific phylogenetic trees and single nucleotide polymorphism analysis. J Virol 2009; 83(5): 2285-97.
[http://dx.doi.org/10.1128/JVI.02180-08] [PMID: 19109389] ]. The International Committee on Taxonomy of Viruses currently classifies polyomaviruses primarily according to the sequence identity of their LTag genes. This system has been questioned by phylogenetic studies suggesting that the evolutionary history of LTag and major capsid protein VP1 is divergent and that some modern polyomaviruses represent chimeric lineages [18Yu J, Boyapati A, Rundell K. Critical role for SV40 small-t antigen in human cell transformation. Virology 2001; 290(2): 192-8.
[http://dx.doi.org/10.1006/viro.2001.1204] [PMID: 11883184] , 19Maurizio P, Etienne XK. The potential therapeutic usefulness of targeting BK Polyomavirusin prostate cancer. Chemo Open Access 2015; 5: 1-2.]. Based on the serological and genotyping method, the BKV isolated from various locations of the world is categorized into four subtypes [21Kaydani GA, Makvandi M, Samarbafzadeh A, et al. Prevalence and distribution of BK virus subtypes in renal transplant recipients referred to golestan hospital in ahvaz, Iran. Jundishapur J Microbiol 2015; 21(8): e16738.]. Subtype I is the most dominant and has a worldwide distribution, followed by subtype IV which is mostly isolated from East Asia. However, subtypes II and III have low frequencies compared to the others [22Andi K, Olaf R, Bininda E, Peter W, Roland Z. Evolution of four BK virus subtypes, infection, genetics and evolution. Infect Genet Evol 2008; 8: 632-43.
[http://dx.doi.org/10.1016/j.meegid.2008.05.006] ]. In addition, the phylogenic investigations revealed that there were four subgroups of subtype I, subgroups 1/a, 1/b-1, 1/b2 and 1/c. While, Subtype IV is divided into six subgroups, 4/a-1, 4/a-2, 4/b-1, 4/b-2, 4/c-1 and 4/c-2 [21Kaydani GA, Makvandi M, Samarbafzadeh A, et al. Prevalence and distribution of BK virus subtypes in renal transplant recipients referred to golestan hospital in ahvaz, Iran. Jundishapur J Microbiol 2015; 21(8): e16738.].
In Sudan, the relationship between viruses and cancer has been studied in only a limited studies [23Gorish BM. Frequency of HBV among hepatocellular carcinoma patients in khartoum state, sudan. J Carcinog Mutagen 2018; 9: 320., 24Dafalla AB, Elnil YF, Gorish BM. Seroprevalence of cytomegalovirus infection among leukemic patients in khartoum state. Virol Mycol 2018; 7: 183.]. In our previous study, the existence of the relationship between BKV and prostate cancer was determined [25Gorish BMT, Ournasseir MEH, Shammat IM. A correlation study of BK Polyoma Virus infection and prostate cancer among sudanese patients - immunofluorescence and molecular based case-control study. Infect Agent Cancer 2019; 14: 25.
[http://dx.doi.org/10.1186/s13027-019-0244-7] [PMID: 31548852] ] and the molecular characterization of BKV was described because only a few researchers have addressed BKV Subtypes.However, most previous studies have focused only on the isolates related to BKV nephropathy. The effect of each subtype on the clinical implications had previously been highlighted by various researchers [26Motazakker M, Bagheri M, Imani M. Subtyping of BK Virus in iranian turkish renal transplant recipients by RFLP-PCR. Maedica (Buchar) 2012; 7(1): 10-3.
[PMID: 23118813] , 27Wang ZY, Hong WL, Zhu ZH, et al. Phylogenetic reconstruction and polymorphism analysis of BK virus VP2 gene isolated from renal transplant recipients in China. Exp Ther Med 2015; 10(5): 1759-67.
[http://dx.doi.org/10.3892/etm.2015.2723] [PMID: 26640547] ]. Some researchers have reported that the characterization of the genetic mutations of BK virus ge may have biological and clinical effects [28Sharma PM, Gupta G, Vats A, Shapiro R, Randhawa P. Phylogenetic analysis of polyomavirus BK sequences. J Virol 2006; 80(18): 8869-79.
[http://dx.doi.org/10.1128/JVI.00510-06] [PMID: 16940499] ]. Thus, our study attempted to determine the phylogenetic characterization of BKV strains among Sudanese patients with a prostate disorder, such as prostate cancer and benign prostatic hyperplasia.
2. MATERIALS AND METHODS
2.1. Overall Design
A cross-sectional study was carried out based on the samples from our previous work on BKV to determine the phylogenetic characterization of BKV strains among Sudanese patients with a prostate disorder, such as prostate cancer and benign prostatic hyperplasia.
2.2. Study Population
A total of 8 samples from our previous work on BKV were studied in this investigation [25Gorish BMT, Ournasseir MEH, Shammat IM. A correlation study of BK Polyoma Virus infection and prostate cancer among sudanese patients - immunofluorescence and molecular based case-control study. Infect Agent Cancer 2019; 14: 25.
[http://dx.doi.org/10.1186/s13027-019-0244-7] [PMID: 31548852] ]. These samples were randomly selected from 21 positive samples. Samples 1 to 5 were obtained from PCa patients, while samples 6 and 7 were taken from BPH patients as well as one urine sample from a patient with BPH. All the patients were referred to central hospitals in Khartoum. Each test was done considering one sample as one patient.
2.3. Data Collection
An interviewer administered questionnaire was used to ask participants about their demographic, socioeconomic, and geographical afflation, cadmium contact, alcohol consumption, cancer grade along with detailed contact information for the communication of result and subsequent treatment if necessary.
2.4. Ethical Statement
Before conducting the study, the proposal of the study was ethically approved by the ethical committee of Omdurman Islamic University and the Ministry of Health. After that, informed consent from each patient and permission from the general managers of the hospitals were obtained.
2.5. Large T Antigen Gene as a Target for BKV Molecular Characterization
The first genotyping schema for BKV described by Jin et al. in 1993 was based on a very short segment of the VP1 gene (nucleotides 1744 to 1812). While phylogenetic trees based on the LTA sequences were introduced and validated recently [20Luo C, Bueno M, Kant J, Martinson J, Randhawa P. Genotyping schemes for polyomavirus BK, using gene-specific phylogenetic trees and single nucleotide polymorphism analysis. J Virol 2009; 83(5): 2285-97.
[http://dx.doi.org/10.1128/JVI.02180-08] [PMID: 19109389] ]. The International Committee on Taxonomy of Viruses currently classifies polyomaviruses primarily according to the sequence identity of their LTag genes. This system has been questioned by phylogenetic studies suggesting that the evolutionary history of LTag and major capsid protein VP1 is divergent and that some modern polyomaviruses represent chimeric lineages [18Yu J, Boyapati A, Rundell K. Critical role for SV40 small-t antigen in human cell transformation. Virology 2001; 290(2): 192-8.
[http://dx.doi.org/10.1006/viro.2001.1204] [PMID: 11883184] , 19Maurizio P, Etienne XK. The potential therapeutic usefulness of targeting BK Polyomavirusin prostate cancer. Chemo Open Access 2015; 5: 1-2.]. Therefore, the standard BKV subtyping and subgrouping was based on the phylogenetic analysis of 170 fragments within VP1 and Large T antigen genes (nucleotides 1564 to 2215 and 171 3021 to 3715, respectively) [29Descamps V, Martin E, Morel V, et al. Comparative evaluation of three nucleic acid-based assays for BK virus quantification. J Clin Microbiol 2015; 53(12): 3822-7.
[http://dx.doi.org/10.1128/JCM.02116-15] [PMID: 26424842] , 30Ibrahim AH, Hisham NA, Yousif FH, et al. Molecular characterization of polyomaviruses (BKV, JCV) in a symptomatic kidney transplant recipients in sudan. AJIDM 2016; 4: 44-51.].
However, in our study, only the Large T antigen gene amplification product (176 bp) is utilized in order to confirm the BKV isolates and to construct a simple phylogenetic tree that will give an idea about the subtypes which circulate in Sudanese patient with a prostate disorder, such as prostate cancer and benign prostatic hyperplasia. Unfortunately, subgroups of the BKV subtypes were not recognized in this study as the study was based only on the LTAg gene to conduct the phylogenetic analysis. The BKV subgrouping will be a very interesting topic for future studies.
2.6. Viral DNA Extraction from Tissue Biopsy and Urine Specimens
About 25 mg of each tissue sample was incubated in lysis buffer (20 µl proteinase K (200 mg/ml) and 5µl of RNAase A). The DNA extraction was performed by the DNeasy® Tissue Kit (QIAGEN Company) according to the manufacturer’s instructions and stored at -80 ºC until further analysis. The quality of DNA was checked using the Nano-drop test. On the other hand, one ml of urine sample was incubated in lysis buffer (20 µl proteinase K (200 mg/ml) and 5µl of RNAase A). The DNA extraction was performed by the DNeasy® Urine Kit (QIAGEN Company) according to the manufacturer’s instructions and stored at -80ºC until further analysis.
2.7. Target Large T Antigen Gene Amplification
BK viral early gene region (Large T antigen region176bp) was amplified in a master mix reaction with a volume of 25μl containing 5 μl of DNA sample and 1µl of specific forward and reverse primer for primer F: 5′-AGTCTTTAGGGTCTT CTAC C-3′ and BK127-R: 5′-GGTGCCAACCTATGGA ACAG-3′ [30Ibrahim AH, Hisham NA, Yousif FH, et al. Molecular characterization of polyomaviruses (BKV, JCV) in a symptomatic kidney transplant recipients in sudan. AJIDM 2016; 4: 44-51.]. and was completed to 25µl with 13 µl distilled water. The amplification was done by using a TC-3000 conventional PCR Thermal Cycler (USA), completing 40 cycles of denaturation at 94°C for 1 min followed by initial denaturation at 94°C for 5 min, annealing at 55°C for 1 min, extinction at 72°C for 2 min and a final extension at the same temperature for 5 min. Finally, the conventional PCR products were subjected to gel electrophoresis (2% agarose gels) to visualize the band of the amplified target gene region with ethidium bromide (0.5 μg/mL) for 30 min in a UV-gel documentation system. The purified BKV genome of the Dunlop strain was used as a positive control, while distilled water was used as a negative control in each PCR run. The primer design and PCR protocol were implemented following the procedure used by Ibrahim et al. in 2016 [30Ibrahim AH, Hisham NA, Yousif FH, et al. Molecular characterization of polyomaviruses (BKV, JCV) in a symptomatic kidney transplant recipients in sudan. AJIDM 2016; 4: 44-51.].
2.8. Purification of the DNA Product Before the Sequencing Process
The success of automated sequencing critically depends on having a high purity template in the correct concentration. Therefore, the PCR products (Fig. 2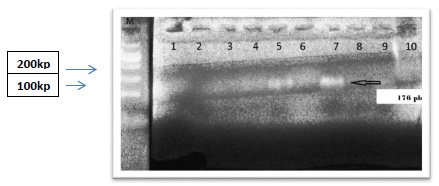 ) were purified by using a Gel Extraction Kit to remove all the unwanted elements from our template. This step was preceded by the detection of PCR template on a gel to confirm the presence of a specific product with an accurate amount. DNA purification was performed according to the standard guidelines of the Macrogen PCR product purification kit (supplied Macrogen, Inc. Korea).
) were purified by using a Gel Extraction Kit to remove all the unwanted elements from our template. This step was preceded by the detection of PCR template on a gel to confirm the presence of a specific product with an accurate amount. DNA purification was performed according to the standard guidelines of the Macrogen PCR product purification kit (supplied Macrogen, Inc. Korea).
2.9. Sequencing Process
All the purified DNA amplicons (176 bp) (Fig. 2 ), including the positive control, were sent to Macrogen company (Macrogen, Inc. Korea) for direct sequencing with 10 pmol of related PCR primers bi-directionally.
), including the positive control, were sent to Macrogen company (Macrogen, Inc. Korea) for direct sequencing with 10 pmol of related PCR primers bi-directionally.
2.10. Bioinformatics Analysis
The obtained nucleotide sequences of large T antigen genes were assembled manually by the Finch TV software, and the cleaned sequences were searched for sequence similarity using nucleotide BLAST (http://blast.ncbi.nlm.nih.gov/ Blast.cgi) and Sequence Similarity Search - BLAST - GenomeNet. Highly similar sequences were retrieved from NCBI and subjected to multiple sequence alignment using the BioEdit. Protein translation and modeling was done online by the CPH model server-3.2 and the EXPASSY translate tools, respectively. The phylogenetic tree was generated by a maximum likelihood method using the MEGA6 program from the aligned nucleotide sequences. The statistical robustness and reliability of the branching order within each phylogenetic tree were confirmed by applying bootstrap resampling (n = 1000 replicates). Sequences from different countries were retrieved from the gene bank and included in the generation of phylogenetic trees.
2.11. Quality Control
To exclude that point mutation is not an error generated during the sequencing process and to confirm the stability of the reagent, we included a purified BKV genome of the Dunlop strain as a positive control and distilled water as a negative control. The positive control yielded a sequence of nucleotides that were identical to that of the gene bank BKV-DUN (JO2038). Briefly, the amplification of beta-globin was done for all the samples to control DNA extraction. All the experiments were carried out in a contamination-free environment.
3. RESULTS
3.1. Patients Description
A total of 8 samples from our previous work on the BKV were studied in this investigation. Samples 1 to 5 were obtained from PCa patients, while samples 6 and 7 were taken from BPH patients as well as one urine sample from a patient with BPH. Patients were aged between 62 and 74 years with an average age of 68 ± 2.4 years. All the patients included were those who were referred to the central hospitals in Khartoum, three were from Central Sudan and two from the Western part. In addition, two patients were from the Northern parts and one from Southern Sudan. The histological identification Gleason scores for samples 1, 2, 3, 4 and 5 were 7, 6, 8, 7 and 8, respectively (Tables 1 and 2).
3.2. Blasting Result
BLAST nucleotide search revealed that isolates 1, 3 and 4 had a 100% nucleotide identity with some of the band gaps with BKV isolates from Iran, Japan, Germany, USA and Zambia, having a GenBank accession number of GU130546.1, LC185075.1, NC_025892, CAP-m9-USA-AY628233, and NC_025896, respectively (Table 3). While the rest of the isolates had a 99% degree of similarity in the nucleotides with the USA isolate, which had a GenBank accession number of NC_001538 (Table 4). Multiple sequence alignment of translated protein showed that isolates 1, 3 and 4 had identical amino acids with Iran, Japan, Germany, USA and Zambia, which had a GenBank accession number of GU130546.1, LC185075.1, NC_025892, CAP-m9-USA-AY628233 and NC_025896, respectively (Tables 3 and 5). Moreover, the result identified a silent mutation within the nucleotide sequences of isolates 7, 6 and 2 at positions 262, 264 and 261, respectively. Once the type of mutation was silent, these isolates (2,6 and 7) were considered as identical amino acid sequences with the USA BKV isolate (GenBank accession number NC_001538) (Tables 4 and 6). The most remarkable result that emerged from our data was the detection of a point transversion mutation in the sequences of isolates 5 and 8, where nucleotide adenine (A) was replaced by Cytosine (C) at position 276 which changed the codon AAC to ACC resulting in the replacement of amino acid Asparagine (N) by Threonine (T) at position 253 (Tables 4 and 6).

Multiple nucleotide sequence alignments. The alignments were performed using the Clustal W2 sequence alignment method through MEGA X program. The table showed the identical nucleotide sequence alignment between BKV isolates 1,3,4 of prostate cancer and isolates from different country in the world with the presence of some gaps.

Multiple nucleotide sequence alignments. The alignments were performed using the Clustal W2 sequence alignment method through MEGA X program. The table describe 99% nucleotides similarity between isolates 2,5,6,7,8 and USA isolate with some point mutations that either to be silent which can be exemplified by isolate 7,6 and 2 in position 262,264 and 261 respectively or transversion mutation in isolates 5,8 which reflect a mutation in position 276 that result in Amino acid Substitution.

Multiple sequence amino acids alignments. The alignments was done by using Clustal W2 sequence alignment method through MEGA X software. isolates 1,3 and 4 were showed an identical amino acids similarity with isolates from IRAN, JAPAN, GERMANY, USA and ZAMBIA with Gen Bank accession number of GU130546.1, LC185075.1, NC_025892, CAP-m9-USA-AY628233 and NC_025896 respectively.

Multiple sequence amino acids alignments. The alignments was done by using Clustal W2 sequence alignment method through MEGA X software. a transvertion mutation was observed in position 253 for isolate 5 and 8 when compared to USA 001538 isolate sequences. While isolate 2, 6 and 7 have similar amino acids sequence similarity with USA 001538 isolate.
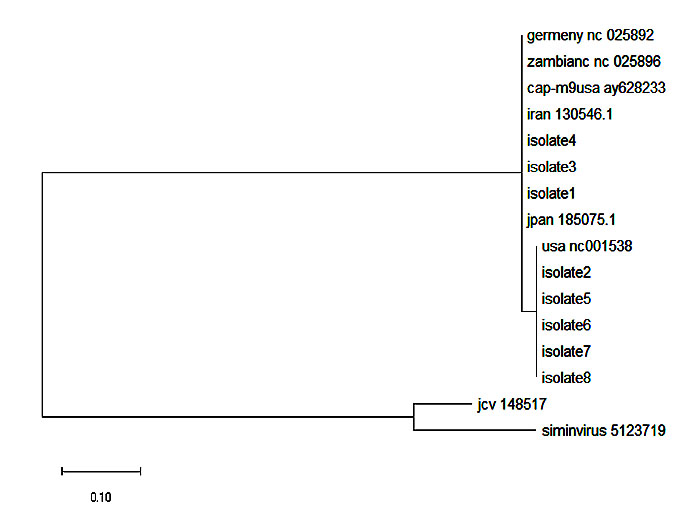 |
Fig. (3) Phylogenetic analysis of the BKV Large T antigen gene. |
Another notable finding in our study was that BKV isolate 5 was obtained from the tissue sample of a PCa patient from Southern Sudan. While BKV isolate 8 was amplified from the urine sample of a BPH patient from Western Sudan. Interestingly, cadmium contact and alcohol consumption were observed for these patients, and it was found that the first patient did not have cadmium contact and did not consume alcohol, while the second was only in contact with cadmium (Table 2).
3.3. Phylogenetic Analysis Result
Phylogenetic analysis of the sequences recovered from the GenBank as well as the sequences obtained from the patient samples was performed. Complete sequences retrieved from the Gen Bank were aligned. A phylogenetic tree was then constructed in order to assemble all the 8 isolates according to the BKV subtypes and subgroups. The results showed that all the Sudanese BKV isolates belonged to the BKV Subtype-1 and were arranged into two subgroups; three isolates were combined with the same clade of GU130546.1, LC185075.1, NC_025892, CAP-m9-USA-AY628233 and NC_025896. While the rest of the isolates were arranged together with the same clade with the USA isolate Gen Bank accession number NC_001538 (Fig. 3 ). These data reflected the distribution of BKV genotypes observed in the worldwide population, suggesting that this cohort of reference strains are relevant.
). These data reflected the distribution of BKV genotypes observed in the worldwide population, suggesting that this cohort of reference strains are relevant.
4. DISCUSSION
This study is not the first report on the molecular characterization of BKV in Sudan. However, most of the previous works have focused solely on the isolates associated with BKV nephropathy. Therefore, our study is the first of its kind to address the molecular characterization of BKV for PCa patients. Historically, BKV subtyping and subgrouping were based mainly on the phylogenetic analysis of some partial gene sequences (vp1 gene). While phylogenetic trees based on LTA sequences have been recently introduced and validated [20Luo C, Bueno M, Kant J, Martinson J, Randhawa P. Genotyping schemes for polyomavirus BK, using gene-specific phylogenetic trees and single nucleotide polymorphism analysis. J Virol 2009; 83(5): 2285-97.
[http://dx.doi.org/10.1128/JVI.02180-08] [PMID: 19109389] ]. This study was based only on the large T antigen gene product 176 bp to construct the phylogenetic tree. All the sequences of the samples were analyzed using various bioinformatics tools in order to understand genotype variability, molecular epidemiology and evolutionary adaptability of circulating strains. The purpose for focusing on the LTAg gene in our study was to determine the correlation between the BKV and prostate cancer in our previous work and to find a positive correlation between the presence of the virus and PCa and it is well known that LTAg promotes cellular transformation by interfering with the tumor suppressor functions of p53 [4Delbue S, Matei DV, Carloni C, et al. Evidence supporting the association of polyomavirus BK genome with prostate cancer. Med Microbiol Immunol (Berl) 2013; 202(6): 425-30.
[http://dx.doi.org/10.1007/s00430-013-0304-3] [PMID: 23821367] -6Provenzano M, Keller P. The potential therapeutic usefulness of targeting BK polyoma virus in prostate cancer. Chemo Open Access 2015; 5: 1.]. Thus, in this study, a point mutation was determined that could arise in this gene, resulting in antigen change. The latter event may affect the effectiveness of the antigen to bind with the antigen suppressing the tumor; this effect can be positive or negative.
In this study, there were two remarkable findings. Firstly, 3 out of 5 PCa BKV isolates were clustered into the same clade with Iranian and Japanese isolates. A previous study in Sudan revealed the BKV LTAg sequences among kidney transplant recipients and reported that three of these isolates had nucleotide similarities with the Japanese isolates. However, their study differed from our study in the clinical source of the isolates. They obtained their isolates from the urine samples of kidney transplant patients while our isolates were originated from the tissue samples of the patients with a prostate disorder. Moreover, in Africa, our isolates have similar nucleotides sequences with Zambian BKV isolate, while Ibrahim et al. found a nucleotide similarity between their isolate and the Ethiopian isolate [30Ibrahim AH, Hisham NA, Yousif FH, et al. Molecular characterization of polyomaviruses (BKV, JCV) in a symptomatic kidney transplant recipients in sudan. AJIDM 2016; 4: 44-51.]. Another significant point was that among our studied isolates, 5 BKV isolates (2 from PCa patients, 2 from BPH patients and 1 from the urine of BPH patients) were clustered together in the same clade with the USA strain, however, the similarity was 99%. Having 5 isolates in the same clade despite controlling the PCR condition and preventing DNA contamination can be considered as a strong evidence that the Sudanese isolates are likely to be closer to each other and could show genetic characteristics different from other strains found in the gene bank. Another point which may support our statement is the presence of point mutation which appears to be silent in some isolates, such as 2, 7 and 6 or transversion mutation like isolates 5 and 8. This latest finding may need further investigation considering the fact that both the mutant BKV isolates were from the hospitals in South and West Sudan which are closely related regions.
The histological identification Gleason scores for prostate cancer samples 1, 2, 3, 4 and 5 were 7, 6, 8, 7 and 8, respectively. Only isolate 5, which had a Gleason score of 8, showed a mismatched mutation, while other scores showed identical sequences upon alignment except for isolate 2 in which there was a silent mutation that had no significant impact on the translated amino acids. The association of the transversion mutation with the highest Gleason score in this study could raises a question whether the mutant variant of the BKV leads to a more aggressive type of PCa or not? In order to answer this question, further studies should be conducted by the inclusion of more BKV isolates from patients with prostate cancer grades.
Our result documented the circulation of one subtype (BKV Subtype 1) among Sudanese PCa and BPH patients with different age and geographical affiliations, regardless of the prostate Cancer Gleason scores. This finding was similar to that obtained by Maryam et al. in 2018 who concluded that all the BKV strains which were included in their study were classified under subtype 1. This is not surprising because most previous studies have revealed that despite different clinical samples, subtype I predominates in all the geographical regions of the world with higher frequencies, while subtype IV occurs at lower rates; and subtypes II and III occur rarely [31Chehadeh W, Kurien SS, Nampoory MR. Molecular characterization of BK and JC viruses circulating among potential kidney donors in kuwait. BioMed Res Int 2013; 2013683464
[http://dx.doi.org/10.1155/2013/683464] [PMID: 23936831] -36Ambalathingal GR, Francis RS, Smyth MJ, Smith C, Khanna R. BK polyomavirus: clinical aspects, immune regulation, and emerging therapies. Clin Microbiol Rev 2017; 30(2): 503-28.
[http://dx.doi.org/10.1128/CMR.00074-16] [PMID: 28298471] ]. Unfortunately, as only the LTAg gene was used to conduct the phylogenetic analysis, the BKV subtype 1 subgroups were not determined. The subgrouping of the BKV subtypes will be a very interesting topic for future studies.
In Sudan, there were no phylogenetic analysis reports on the BKV genome detected among patients with PCa and BPH tissues. In our study, more than 65% and 37% of studied samples were derived from PCa and BPH patients. Studies on the diversity of large T antigen gene sequences may have an implicit role in understanding the development of PCa by BKV because the LTAg gene, together with a small T antigen gene, produces antigens that are responsible for virus carcinogenesis [18Yu J, Boyapati A, Rundell K. Critical role for SV40 small-t antigen in human cell transformation. Virology 2001; 290(2): 192-8.
[http://dx.doi.org/10.1006/viro.2001.1204] [PMID: 11883184] , 19Maurizio P, Etienne XK. The potential therapeutic usefulness of targeting BK Polyomavirusin prostate cancer. Chemo Open Access 2015; 5: 1-2.]. Our study had some limitations. Due to a lack of funding, only LTAg gene sequences were used in this study. A few numbers of isolates were included in this study. Therefore, the results may not be generalized. Nevertheless, two new transversion mutations among all the studied patients were consistently reported. This latest finding has not been reported before among Sudanese patients, which increases the value of our findings.
CONCLUSION
The results of this study emphasized the importance of early detection and characterization of newly emerging genotypes. It was concluded that all the BKV isolates which circulate among Sudanese prostate tumoral patients belonged to subtype 1, andtwo isolates showed transversion mutations within the large T antigen amino acids sequence (positions 253), in addition to the presence of a silent mutation in some other isolates. These findings highlighted the need for the molecular detection and typing of BKV in Sudan for a better demonstration of the relationship between the virus and PCa. Moreover, our sequence analysis was done on the Large T antigen gene rather than the VP1 gene which is used as a standard target for the determination of BKV subtypes. Therefore, further studies must be conducted on molecular genotyping of BKV in Sudan by studying phylogeny through the amplification of both the above-mentioned genes with the inclusion of more BKV isolates,focusing on prostate cancer grades and patient age in addition to their geographical affiliations as well as the performance of a cell culture, transfection and BKV rescue to demonstrate that DNA BKV belonged to initially replicating BKV viruses. These may enable a better demonstration of the correlation between the BKV isolates and PCa.
LIST OF ABBREVIATIONS
| PCa | = Prostate Cancer |
| BPH | = Benign Prostatic Hyperplasia |
| LTAg | = Large Tumor Antigen |
| BKV | = BK Virus |
| PCR | = Polemrase Chain Reaction |
AUTHORS' CONTRIBUTION
BG and MO performed the main experiments, BG and MO collected patients’ samples and information. BG and IS designed the experiments and wrote the manuscript. All the authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was ethically approved by the ethical committee of Omdurman Islamic University and the Ministry of Health under reference no. 207MLS/2017, Sudan.
HUMAN AND ANIMAL RIGHTS
No animals were used in this research. All human research procedures followed were in accordance with the guidelines of the World Medical Association Declaration of Helsinki Ethical Principles for Medical Research Involving Human Subjects 1964.
CONSENT FOR PUBLICATION
Informed consent from each patient and permission from the general managers of the hospitals were obtained.
AVAILABILITY OF DATA AND MATERIAL
The datasets used and/or analyzed during the current study are available from the corresponding author (Babbiker M.T. Gorish) on reasonable request.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Miss EnasAbdelkafi who contributed significantly to the collection of the specimen. The author are also grateful to the research lab staff of the veterinary college at the Bahri University, namely Prof. Hind ALnasry, Dr. Mona Khair, Dr. Sufyan Kamal and Miss HalaAbdulgader, for their advice in the molecular analyses. Also, the authors are grateful to all the patients who participated in this study, with our wishes for them to cure very soon.
REFERENCES
| [1] | Verhamme KM, Dieleman JP, Bleumink GS, et al. Triumph Pan European Expert Panel. Incidence and prevalence of lower urinary tract symptoms suggestive of benign prostatic hyperplasia in primary care--the Triumph project. Eur Urol 2002; 42(4): 323-8. [http://dx.doi.org/10.1016/S0302-2838(02)00354-8] [PMID: 12361895] |
| [2] | Grossman DC, Curry SJ, Owens DK, et al. US Preventive Services Task Force. Screening for Prostate Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2018; 319(18): 1901-13. [http://dx.doi.org/10.1001/jama.2018.3710] [PMID: 29801017] |
| [3] | Rodríguez H, Levican J, Muñoz JP, et al. Viral infections in prostate carcinomas in Chilean patients. Infect Agent Cancer 2015; 10: 27. [http://dx.doi.org/10.1186/s13027-015-0024-y] [PMID: 26330890] |
| [4] | Delbue S, Matei DV, Carloni C, et al. Evidence supporting the association of polyomavirus BK genome with prostate cancer. Med Microbiol Immunol (Berl) 2013; 202(6): 425-30. [http://dx.doi.org/10.1007/s00430-013-0304-3] [PMID: 23821367] |
| [5] | Elimam ME, Abdelraof S. Incidence of carcinoma of the prostate in patients with normal prostatic specific antigen following prostatectomy. Glob J Medi Res 2013; 13: 26-35. |
| [6] | Provenzano M, Keller P. The potential therapeutic usefulness of targeting BK polyoma virus in prostate cancer. Chemo Open Access 2015; 5: 1. |
| [7] | Martinez-Fierro ML, Leach RJ, Gomez-Guerra LS, et al. Identification of viral infections in the prostate and evaluation of their association with cancer. BMC Cancer 2010; 10: 326. [http://dx.doi.org/10.1186/1471-2407-10-326] [PMID: 20576103] |
| [8] | Wright JL, Lin DW, Stanford JL. Circumcision and the risk of prostate cancer. Cancer 2012; 118(18): 4437-43. [http://dx.doi.org/10.1002/cncr.26653] [PMID: 22411189] |
| [9] | De Marzo AM, Platz EA, Sutcliffe S, et al. Inflammation in prostate carcinogenesis. Nat Rev Cancer 2007; 7(4): 256-69. [http://dx.doi.org/10.1038/nrc2090] [PMID: 17384581] |
| [10] | Das D, Wojno K, Imperiale MJ. BK virus as a cofactor in the etiology of prostate cancer in its early stages. J Virol 2008; 82(6): 2705-14. [http://dx.doi.org/10.1128/JVI.02461-07] [PMID: 18160432] |
| [11] | Sina A. Detection of Polyomaviral large T antigen in benign prostatic hyperplasia and prostate carcinoma tissues. JKIMSU 2015; 4: 26-33. |
| [12] | Monini P, Rotola A, Di Luca D, et al. DNA rearrangements impairing BK virus productive infection in urinary tract tumors. Virology 1995; 214(1): 273-9. [http://dx.doi.org/10.1006/viro.1995.9928] [PMID: 8525628] |
| [13] | Zambrano A, Kalantari M, Simoneau A, Jensen JL, Villarreal LP. Detection of human polyomaviruses and papillomaviruses in prostatic tissue reveals the prostate as a habitat for multiple viral infections. Prostate 2002; 53(4): 263-76. [http://dx.doi.org/10.1002/pros.10157] [PMID: 12430138] |
| [14] | Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet 1971; 1(7712): 1253-7. [http://dx.doi.org/10.1016/S0140-6736(71)91776-4] [PMID: 4104714] |
| [15] | Tognon M, Corallini A, Martini F, Negrini M, Barbanti-Brodano G. Oncogenic transformation by BK virus and association with human tumors. Oncogene 2003; 22(33): 5192-200. [http://dx.doi.org/10.1038/sj.onc.1206550] [PMID: 12910256] |
| [16] | Dalianis T, Hirsch HH. Human polyomaviruses in disease and cancer. Virology 2013; 437(2): 63-72. [http://dx.doi.org/10.1016/j.virol.2012.12.015] [PMID: 23357733] |
| [17] | Ahuja D, Sáenz-Robles MT, Pipas JM. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation. Oncogene 2005; 24(52): 7729-45. [http://dx.doi.org/10.1038/sj.onc.1209046] [PMID: 16299533] |
| [18] | Yu J, Boyapati A, Rundell K. Critical role for SV40 small-t antigen in human cell transformation. Virology 2001; 290(2): 192-8. [http://dx.doi.org/10.1006/viro.2001.1204] [PMID: 11883184] |
| [19] | Maurizio P, Etienne XK. The potential therapeutic usefulness of targeting BK Polyomavirusin prostate cancer. Chemo Open Access 2015; 5: 1-2. |
| [20] | Luo C, Bueno M, Kant J, Martinson J, Randhawa P. Genotyping schemes for polyomavirus BK, using gene-specific phylogenetic trees and single nucleotide polymorphism analysis. J Virol 2009; 83(5): 2285-97. [http://dx.doi.org/10.1128/JVI.02180-08] [PMID: 19109389] |
| [21] | Kaydani GA, Makvandi M, Samarbafzadeh A, et al. Prevalence and distribution of BK virus subtypes in renal transplant recipients referred to golestan hospital in ahvaz, Iran. Jundishapur J Microbiol 2015; 21(8): e16738. |
| [22] | Andi K, Olaf R, Bininda E, Peter W, Roland Z. Evolution of four BK virus subtypes, infection, genetics and evolution. Infect Genet Evol 2008; 8: 632-43. [http://dx.doi.org/10.1016/j.meegid.2008.05.006] |
| [23] | Gorish BM. Frequency of HBV among hepatocellular carcinoma patients in khartoum state, sudan. J Carcinog Mutagen 2018; 9: 320. |
| [24] | Dafalla AB, Elnil YF, Gorish BM. Seroprevalence of cytomegalovirus infection among leukemic patients in khartoum state. Virol Mycol 2018; 7: 183. |
| [25] | Gorish BMT, Ournasseir MEH, Shammat IM. A correlation study of BK Polyoma Virus infection and prostate cancer among sudanese patients - immunofluorescence and molecular based case-control study. Infect Agent Cancer 2019; 14: 25. [http://dx.doi.org/10.1186/s13027-019-0244-7] [PMID: 31548852] |
| [26] | Motazakker M, Bagheri M, Imani M. Subtyping of BK Virus in iranian turkish renal transplant recipients by RFLP-PCR. Maedica (Buchar) 2012; 7(1): 10-3. [PMID: 23118813] |
| [27] | Wang ZY, Hong WL, Zhu ZH, et al. Phylogenetic reconstruction and polymorphism analysis of BK virus VP2 gene isolated from renal transplant recipients in China. Exp Ther Med 2015; 10(5): 1759-67. [http://dx.doi.org/10.3892/etm.2015.2723] [PMID: 26640547] |
| [28] | Sharma PM, Gupta G, Vats A, Shapiro R, Randhawa P. Phylogenetic analysis of polyomavirus BK sequences. J Virol 2006; 80(18): 8869-79. [http://dx.doi.org/10.1128/JVI.00510-06] [PMID: 16940499] |
| [29] | Descamps V, Martin E, Morel V, et al. Comparative evaluation of three nucleic acid-based assays for BK virus quantification. J Clin Microbiol 2015; 53(12): 3822-7. [http://dx.doi.org/10.1128/JCM.02116-15] [PMID: 26424842] |
| [30] | Ibrahim AH, Hisham NA, Yousif FH, et al. Molecular characterization of polyomaviruses (BKV, JCV) in a symptomatic kidney transplant recipients in sudan. AJIDM 2016; 4: 44-51. |
| [31] | Chehadeh W, Kurien SS, Nampoory MR. Molecular characterization of BK and JC viruses circulating among potential kidney donors in kuwait. BioMed Res Int 2013; 2013683464 [http://dx.doi.org/10.1155/2013/683464] [PMID: 23936831] |
| [32] | Vaezjalali M, Azimi H, Hosseini SM, Taghavi A, Goudarzi H. Different strains of BK polyomavirus: VP1 sequences in a group of iranian prostate cancer patients. Urol J 2018; 15(2): 44-8. [PMID: 29277885] |
| [33] | Krumbholz A, Zell R, Egerer R, et al. Prevalence of BK virus subtype I in Germany. J Med Virol 2006; 78(12): 1588-98. [http://dx.doi.org/10.1002/jmv.20743] [PMID: 17063524] |
| [34] | Jin L, Gibson PE, Knowles WA, Clewley JP. BK virus antigenic variants: Sequence analysis within the capsid VP1 epitope. J Med Virol 1993; 39(1): 50-6. [http://dx.doi.org/10.1002/jmv.1890390110] [PMID: 7678637] |
| [35] | Agostini HT, Brubaker GR, Shao J, et al. BK virus and a new type of JC virus excreted by HIV-1 positive patients in rural Tanzania. Arch Virol 1995; 140(11): 1919-34. [http://dx.doi.org/10.1007/BF01322682] [PMID: 7503691] |
| [36] | Ambalathingal GR, Francis RS, Smyth MJ, Smith C, Khanna R. BK polyomavirus: clinical aspects, immune regulation, and emerging therapies. Clin Microbiol Rev 2017; 30(2): 503-28. [http://dx.doi.org/10.1128/CMR.00074-16] [PMID: 28298471] |