- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
In Vivo Interaction of the Hepatitis Delta Virus Small Antigen with the ELAV-Like Protein HuR
Ana Casaca1, Margarida Fardilha2, Edgar da Cruz e Silva2, Celso Cunha*, 1
Abstract
The small and large delta antigens (S-HDAg and L-HDAg, respectively) represent two forms of the only protein encoded by the hepatitis delta virus (HDV) RNA genome. Consequently, HDV relies, at a large extent, on the host cell machinery for replication and transcription. Until now, only a limited number of cellular proteins were identified as S-HDAg or L-HDAg partners being involved in the modulation of the virus life cycle. In an attempt to identify cellular S-HDAg-binding proteins we made use of a yeast two-hybrid approach to screen a human liver cDNA library. We were able to identify HuR, a ubiquitously expressed protein involved in RNA stabilization, as an S-HDAg partner both in vitro and in vivo. HuR was found to be overexpressed and colocalize with HDAg in human hepatoma cells. siRNA knockdown of HuR mRNA resulted in inhibition of S-HDAg and L-HDAg expression.
Article Information
Identifiers and Pagination:
Year: 2011Volume: 5
First Page: 12
Last Page: 21
Publisher Id: TOVJ-5-12
DOI: 10.2174/1874357901105010012
Article History:
Received Date: 30/7/2010Revision Received Date: 27/10/2010
Acceptance Date: 4/11/2010
Electronic publication date: 24/3/2011
Collection year: 2011
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Unidade de Biologia Molecular, Centro de Malária e outras Doenças Tropicais, Instituto de Higiene e Medicina Tropical, Universidade Nova de Lisboa. Rua da Junqueira 100, 1349-008 Lisboa, Portugal; Tel: +351213652620; Fax: +351213632105; E-mail: ccunha@ihmt.unl.pt
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 30-7-2010 |
Original Manuscript | In Vivo Interaction of the Hepatitis Delta Virus Small Antigen with the ELAV-Like Protein HuR | |
INTRODUCTION
Hepatitis delta virus (HDV) coinfects or superinfects human hepatocytes already infected with hepatitis B virus (HBV) resulting in increased liver damage and risk of cirrhosis and fulminant disease [1Govindarajan S, Chin KP, Redeker AG, Peters RL. Fulminant B viral hepatitis: Role of delta agent Gastroenterology 1984; 6: 1417-20.]. The HDV outer envelope consists of HBV surface antigens (HBsAg) which are necessary for propagation of infection [2Rizzetto M, Hoyer B, Canese MG, Shih JW, Purcell RH, Gerin JL. Delta agent: association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees Proc Natl Acad Sci USA 1980; 77: 6124-8.-4Smedile A, Rizzetto M, Gerin J. Advances in hepatitis D virus biology and disease Prog Liver Dis 1994; 12: 157-75.], and thus HDV is considered to be a HBV satellite virus.
The HDV genome consists of a single-stranded, circular RNA molecule of about 1.7 kilobases (kb) which bears about 70% of internal base-pairing and folds into a rod-like structure similar to plant viroids. The genome contains a single ORF from which two forms of the same protein, the small and large delta antigens (S-HDAg and L-HDAg, respectively), are derived by an RNA editing mechanism catalyzed by cellular adenosine deaminase ADAR1 [5Casey JL, Gerin JL. Hepatitis D virus RNA editing: Specific modification at adenosine in the antigenome RNA J Virol 1995; 69: 7593-600.]. As a consequence, an amber stop codon UAG is transformed into a tryptophan codon UGG extending the ORF by additional 19 aminoacids. Thus, both forms of HDAg share a number of specific domains: a bipartide RNA binding domain [6Lin JH, Chang MF, Baker SC, Govindarajan S, Lai MMC. Characterization of hepatitis delta virus antigen J Virol 1990; 64: 4051-8.], an oligomerization domain [7Lazinski DW, Taylor JM. Relating structure to function in the hepatitis delta virus antigen J Virol 1993; 67: 2677-80.] and a nuclear localization signal [8Alves C, Freitas N, Cunha C. Characterization of the nuclear localization signal of the hepatitis delta virus antigen Virology 2008; 370: 12-21.]. Additionally, L-HDAg was shown to be isoprenylated at a cysteine residue located four aminoacids from the C-terminus [9Glenn JS, Watson JA, Havel CM, White JO. Identification of a prenylation site in the delta virus large antigen Science 1992; 256: 1331-3.]. Despite sequence similarities, different functions have been assigned to these two proteins: S-HDAg is required for nuclear import of HDV RNPs and synthesis of HDV RNA [10Yamaguchi Y, Filipovska J, Yano K, et al. Stimulation of RNA polymerase II elongation by hepatitis delta antigen Science 2001; 293: 124-7.], whereas L-HDAg represses in trans HDV replication and is essential for virion assembly, through interaction between the C-terminal and HBsAg [11Chang FL, Chen PJ, Tu SJ, Wang CJ, Chen DS. The large form of hepatitis antigen is crucial for assembly of hepatitis virus Proc Natl Acad Sci 1991; 88: 8490-4.].
HDV RNA replication involves the redirection of at least cellular DNA-dependent RNA polymerase II. It is believed that HDV RNA synthesis occurs through a rolling-circle mechanism resulting in the formation of multimeric antigenomes which are self-cleaved and ligated at precise monomeric intervals. These antigenomes serve as templates for subsequent synthesis of genomic RNA molecules by a similar mechanism.
HDV is the smallest human pathogen identified so far being highly dependent on host cell factors to accomplish the replication cycle. Several cellular proteins were already reported to bind to HDV RNA and antigens. Some were already shown to play an important role in the virus life cycle but the precise function of a significant number remains elusive (reviewed in [12Greco-Stewart VS, Pelchat M. Interaction of Host Cellular proteins with components of the Hepatitis Delta virus Viruses 2010; 2: 189-212.]). These proteins include ADAR1 which binds to antigenomic RNA during editing [13Wong SK, Lazinski DW. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1 Proc Natl Acad Sci USA 2002; 99: 15118-23.] and RNA polymerase II which was shown to interact with HDV RNA [14Yamaguchi Y, Mura T, Chanarat S, Okamoto S, Handa H. Hepatitis delta antigen binds to the clamp of RNA polymerase II and affects transcriptional fidelity Genes Cells 2007; 12: 863-75.]. Additionally, transcription factors and proteins involved in RNA metabolism, namely the TATA-binding protein [15Abrahem A, Pelchat M. Formation of an RNA polymerase II preinitiation complex on an RNA promoter derived from the hepatitis delta virus RNA genome Nucleic Acids Res 2008; 36: 5201-11.], the zinc finger transcription factor YY1 [16Huang WH, Mai RT, Lee YH. The transcription factor YY1 and its associated acetyltranspherases CBP and p300 interact with hepatitis delta virus antigens and modulate the hepatitis delta virus RNA replication J Virol 2008; 82: 7313-24.], and the polyrimidine tract-binding protein associated splicing factor (PSF; [17Greco-Stewart VS, Thibault CS, Pelchat M. Binding of the polypirimidine tract-binding protein-associated splicing factor (PSF) to the hepatitis delta virus RNA Virology 2006; 356: 35-44.]), were also reported to bind to the virus RNA and antigens. Nucleolar proteins, such as nucleolin [18Lee CH, Chang SC, Chen CJ, Chang MF. The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting J Biol Chem 1998; 273: 7650-6.] and B23 were also shown to bind to S-HDAg to form a large complex which was proposed to enhance HDV replication [19Huang WH, Yung BY, Syu WJ, Lee YH. The nucleolar phosphoprotein B23 interacts with hepatitis delta antigens and modulates hepatitis delta virus RNA replication J Biol Chem 2001; 276: 25166-75.]. Finally, using a yeast two hybrid approach it was possible to identify the nuclear export signal-interacting protein (NESI) as cellular counterpart of L-HDAg being essential to promote nuclear export and assembly of HDV genomic RNA [20Wang YH, Chang SC, Huang C, Li YP, Lee CH, Chang MF. Novel nuclear export signal-interacting protein, NESI, critical for the assembly of hepatitis delta virus J Virol 2005; 79: 8113-20.]. Moreover, proteomic approaches were used to identify changes in cellular protein expression caused by the presence of HDV components [21Mota S, Mendes M, Penque D, Coelho AV, Cunha C. Changes in the proteome of Huh7 cells induced by transient expression of hepatitis D virus RNA and antigens J Proteomics 2008; 71: 71-9., 22Mota S, Mendes M, Freitas N, Penque D, Coelho AV, Cunha C. Proteome analysis of a human liver carcinoma cell line stably expressing hepatitis delta vírus ribonucleoproteins J Proteomics 2009; 72: 616-27.] and a study making use of immunoprecipitation followed by mass spectrometry analysis lead to identification of more than 100 proteins that may interact with S-HDAg [23Cao D, Haussecker D, Huang Y, Kay MA. Combined proteomic-RNAi screen for host factors involved in human hepatitis delta virus replication RNA 2009; 15: 1971-9.].
Despite the increasing number of cellular factors described as HDV RNA or antigens partners, the role of most of these interactions in HDV replication were not elucidated, and many steps of the virus life cycle remain unclear. Furthermore, given the simplicity of the virus and the limited number of host factors found to play a role in its replication, it is highly probable that still a considerable number remains to be uncovered.
In an attempt to identify new cellular partners of S-HDAg, in this work we screened a human liver cDNA library using a yeast two-hybrid approach. The HuR protein, a member of the ELAV (embryonic lethal abnormal vision) family of RNA-binding proteins, was found to specifically interact with S-HDAg in this system.
HuR is a nucleocytoplasmic shuttling protein [24Fan XC, Steitz J. HNS, a nuclear-cytoplasmic shuttling sequence in HuR Proc Natl Acad Sci USA 1998; 95: 15293-8.] ubiquitously expressed in proliferating cells, and was previously reported to bind to AU-rich elements (AREs) of mRNAs affecting their stabilization and expression (reviewed in [25Brennan CM, Steitz JA. HuR and mRNA stability Cell Mol Life Sci 2001; 58: 266-77.]). This ability enables HuR to play a variety of biological roles and to be involved in different cellular mechanisms including apoptosis [26Lal A, Kawai T, Yang X, Mazan-Mamczarz K, Gorospe M. Antiapoptotic function of RNA-binding protein HuR effected through prothymosin alpha EMBO J 2005; 24: 1852-62., 27Mazroui R, Di Marco S, Clair E, et al. Caspase-mediated cleavage of HuR in the cytoplasm contributes to pp32/PHAP-I regulation of apoptosis J Cell Biol 2008; 180: 113-27.], oncogenesis [28Lopez de Silanes I, Lal A, Gorospe M. HuR: posttranscriptional paths to malignancy RNA Biol 2005; 2: 11-3.], and cellular response to stress [29Gorospe M. HuR in the mammalian genotoxic response: post-transcriptional multitasking Cell Cycle 2003; 2: 412-.]. HuR was also found to directly interact with the HIV-1 reverse transcriptase to modulate reverse transcription by a still unknown mechanism [30Lemay J, Mandou-Peindara P, Bader T, et al. HuR interacts with human immunodeficiency virus type I reverse transcriptase, and modulates reverse transcription in infected cells Retrovirology 2008; 5: 47.], to bind to 3’ ends of hepatitis C virus (HCV) RNA [31Spangberg K, Wiklund L, Schwartz S. HuR, a protein implicated in oncogene and growth factor mRNA decay, binds to the 3’ ends of hepatitis C virus RNA of both polarities Virology 2000; 274: 378-90.] and to the 3’ untranslated region of human papillomavirus RNA [32Sokolowski M, Furneaux H, Schwartz S. The inhibitory activity of the AU-rich RNA element in the human papillomavirus type 1 late 3' untranslated region correlates with its affinity for the elav-like HuR protein J Virol 1999; 73: 1080-91.]. Additionally, it was recently reported that HuR is involved in the translational control of HIV-1 and HCV IRES (internal ribosome entry site) elements [33Rivas-Aravena A, Ramdohr P, Vallejos M, et al. The ELAV-like protein HuR exerts translational control of viral internal ribosome entry sites Virology 2009; 392: 178-85.]. The authors also showed that regulation of IRES elements seems not to involve a direct interaction of HuR with the RNA.
Here, we report that HuR and S-HDAg interact both in vivo and in vitro, and colocalize in human liver cells. Furthermore, HuR was found to be overexpressed in cells expressing HDV RNPs. Finally, HuR silencing with specific siRNAs lead to a decrease in both S-HDAg and L-HDAg expression. The obtained results suggest that HuR plays an important role in HDV replication.
MATERIALS AND METHODOLOGY
Yeast Two-Hybrid Screening
The yeast two-hybrid screening was performed in Saccharomyces cerevisiae using the Matchmaker system (BD Biosciences).
The gene coding for the S-HDAg was PCR amplified from pSVL-AgS [11Chang FL, Chen PJ, Tu SJ, Wang CJ, Chen DS. The large form of hepatitis antigen is crucial for assembly of hepatitis virus Proc Natl Acad Sci 1991; 88: 8490-4.], using the primers 5’-ttatcaattgatgag ccggtccgagtcg-3 (forward) and 5´-ttatgtcgacctatggaaatccct ggtttccc-3’ (reverse). The resulting fragment was cleaved (MunI/BamHI) and inserted into pAS2-1 vector (BD Biosciences) in the local EcoRI/BamHI, to obtain the plasmid pAS2-1/S-HDAg. This plasmid contains the S-HDAg entire gene fused to the GAL4 DNA binding domain (BD) and was used as bait to screen a human liver cDNA library representing fusions of the GAL4 activation domain (AD) pretransformed into the Y187 yeast strain.
Screening of the library was performed according to manufacturer’s instructions. To reduce the false positives after selection, the selected yeast colonies were transferred to Trp, Leu, His and Ade dropout nutritional selection media and assayed for MEL1 activity.
Prey plasmids encoding human liver proteins that interacted with S-HDAg in the MEL1 positive yeast clones were isolated after transformation in E. coli DH5α. AD plasmids were recovered from positive yeast clones and retransformed into native AH109 yeast with the bait plasmid pAS2-1/S-HDAg to retest the interactions. Finally, the recovered plasmids were subjected to DNA sequencing.
Production and Purification of Recombinant Proteins
The bacterial expression vectors pGEX-6P-2 (GE HealthCare) and pET28c (Novagen) were used to produce GST or His-tagged fusion proteins, respectively.
The fusion vector for expression of GST/S-HDAg (pGEX-6P-2/S-HDAg) was constructed by digesting plasmid pEGFP-C1-HDAg (Tavanez et al. 2002) using the restrictions enzymes BglII and SalI followed by insertion of the obtained S-HDAg cDNA fragment into the XhoI/BamHI cloning sites of pGEX-6P-2. To construct the plasmid pET28c/HuR for expression of His6/HuR fusion proteins, the HuR cDNA cloned in pCMV_SPORT6 (Image clone # 2901220) was amplified using the following primers: 5’-ttatcaattgccatgtctaatggttatgaagaccacatgg-3’ (forward) and 5’- ttatgtcgaccgagttatttgtgggacttgttgg-3´ (reverse). The resulting fragment was cleaved with MunI and SalI and cloned into pET28c, in the EcoRI/SalI sites.
Resulting recombinant plasmids pGEX-6P-2-HDAg and pET28c/HuR were used to transform E. coli BL21 and E. coli BL21-CodonPlus-(DE3)-RP competent cells (Stratagene), respectively. The expression of both GST/S-HDAg and His6/HuR were induced with 1 mM isopropyl-B-D-thiogalactopyranoside (IPTG) at 37º C for 2 h. Cells were then harvested by centrifugation, washed with phosphate-buffered saline (PBS) and resuspended in PBS containing 0.1mg/mL lysozyme (Sigma). After three freeze-thaw cycles the obtained samples were treated with DNase I (Roche).
For purification of recombinant S-HDAg, protein extracts were loaded onto a GSTrap FF 1mL column (GE Healthcare). After washing, the GST-tag was removed by digesting with the PreScission protease (GE Healthcare) according to the instructions of the manufacturer. Finally, the eluted protein was dialyzed against PBS and quantified using a Bradford assay based kit (BioRad).
Cell Culture
Two different cell lines were used in this study. The human hepatocellular carcinoma cell line (Huh7) and the corresponding HDV cDNA stably transfected cell line (Huh7-D12), which constitutively expresses HDV ribonuc-leoproteins [35Cheng D, Yang A, Thomas H, Monjardino J. Characterization of stable hepatitis delta-expressing cell lines: Effect of HDAg on cell growth Prog Clin Biol Res 1993; 382: 149-53.]. Cells were grown as monolayers in RPMI 1640 medium (Sigma) supplemented with 10% fetal bovine serum (Invitrogen). Cells were cultured at 37ºC in a humidified atmosphere containing 5% CO2.
Blot Overlay
Blot overlay assays were performed essentilally as described [36Browne GJ, Fardilha M, Oxnham SK, et al. Sarp, a new alternatively spliced protein phosphatase 1 and DNA interacting protein Biochem J 2007; 402: 187-96.]. Briefly, increasing amounts of bacterial extracts containing His6/HuR recombinant protein were separated on 12% SDS/PAGE gels. The proteins were subsequently transferred to a nitrocellulose membrane that was then overlaid with 40 µg purified recombinant S-HDAg protein. After washing, bound S-HDAg was detected by immunoblot using a rabbit polyclonal anti-S-HDAg antibody (B3; [37Saldanha J, Homer E, Goldin P, Thomas HC, Monjardino J. Cloning and expression of an immunodominant region of the hepatitis delta antigen J Gen Virol 1990; 71: 471-5.]) followed by incubation with a secondary anti-rabbit IgG antibody conjugated with peroxidise (BioRad). Membrane development was achieved using an enhanced chemiluminescence system (ECL; GE Healthcare) according to the specifications of the manufacturer.
Coimmunoprecipitation
For coimmunoprecipitation, Huh7 or Huh7-D12 cells were collected in lysis buffer [50mM Tris-HCl (pH 7.5), 150mM NaCl, 1% Nonidet P-40 and 10% glycerol] supplemented with complete protease inhibitor cocktail (ROCHE). Cell extracts were precleared with protein G Dynabeads (Invitrogen) and incubated for 2 h at 4ºC with a polyclonal rabbit anti-HDAg antibody (B3; [37Saldanha J, Homer E, Goldin P, Thomas HC, Monjardino J. Cloning and expression of an immunodominant region of the hepatitis delta antigen J Gen Virol 1990; 71: 471-5.]). Protein G Dynabeads (Invitrogen) were then added and immunoprecipitates were washed three times with PBS. Antigen-antibody complexes were recovered by boiling in SDS sample buffer and analyzed by western blot as described below.
Western Blot
To prepare protein samples for western blot, Huh7 or Huh7-D12 cells were first harvested, washed twice with PBS, and pelleted by centrifugation. The cell pellet was re-suspended in SDS sample buffer and boiled. Western blot was performed as previously described [38Cunha C, Monjardino J, Chang D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus in the nucleus of human cells RNA 1998; 4: 680-93.].
Protein samples, either recombinant or immunoprecipitated proteins, were separated by SDS-PAGE on 12% polyacrilamide gels and electroblotted onto nitrocellulose membranes (Schleicher & Schuell). Blots were then incubated for 1 h with one of the following primary antibodies: mouse monoclonal antibody against HuR (Santa Cruz Biotechnology), rabbit polyclonal anti-HDAg antibody B3 [37Saldanha J, Homer E, Goldin P, Thomas HC, Monjardino J. Cloning and expression of an immunodominant region of the hepatitis delta antigen J Gen Virol 1990; 71: 471-5.], and mouse anti-His-tag monoclonal antibody (Sigma-Aldrich). When appropriate the control house-keeping protein clathrin was also detected using an anti-clathrin antibody (Santa Cruz Biotechnology), and used to normalize for the amount of each selected protein. After washing with PBS containing 0.1% Tween 20 and 2% low fat milk powder, blots were further incubated with appropriate secondary antibodies conjugated with horseradish peroxidase (BioRad). Membrane development was performed using an enhanced chemiluminescence system (ECL; GE Healthcare).
When needed the density of the bands was quantified using the ImageJ software (http://rbs.info.nih.gov/ij/).
Immunofluorescence and In Situ Hybridization
Indirect imunofluorescence and in situ hybridization were essentially performed as earlier described [38Cunha C, Monjardino J, Chang D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus in the nucleus of human cells RNA 1998; 4: 680-93.]. In brief, for immunofluorescence, cells grown on coverslips were rinsed briefly in PBS, fixed with 3.7% formaldehyde and permeabilized with 0.5% Triton X-100 in PBS, for 10 min, at room temperature. Cells were then rinsed with PBS containing 0.05% Tween 20 (PBS-T) and incubated with primary antibodies (polyclonal rabbit anti-HDAg serum B3 and/or mouse monoclonal against HuR; Santa Cruz Biotechnology). After washing with PBS-T, cells were incubated with appropriate secondary antibodies conjugated to either FITC or Texas Red (Jackson Immunoresearch Laboratories). Finally, the coverslips were mounted in Vectashield (Vector Laboratories, UK).
In situ hybridization was performed using a digoxigenin-11-dUTP (Roche) nick translated plasmid pSVL(D3) [39Kuo MYP, Chao M, Taylor J. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: Role of delta antigen J Virol 1989; 63: 1945-50.] as probe. This plasmid contains a trimer of full-length genomic HDV cDNA cloned in pSVL (GE Healthcare). After fixation and permeabilization, as described above for immunofluorescence, cells were sequentially treated with 70% formamide at 73ºC followed by incubation with 50% formamide at 73ºC, for 5 min, and finally subjected to hybridization. After hybridization, digoxigenin detection was performed using a monoclonal anti-digoxigenin antibody conjugated with FITC (Roche) followed by incubation with a secondary anti-FITC antibody conjugated with Alexa-488 (Jackson Immuno Research Laboratories). Finally, the coverslips were mounted in Vectashield (Vector Laboratories, UK).
Confocal Microscopy and Image Analysis
Fluorescent-labeled samples were analyzed under a Zeiss LSM 510 META confocal microscope. Green fluorescence was detected using an Argon Ion laser (488 nm) and red fluorescence was detected using a Helium-Neon laser (543 nm). Calibration of the equipment was achieved using multicolor fluorescent beads (Molecular Probes, USA) and a dual-band filter which allows simultaneous visualization of green and red fluorescence.
The quantitative colocalization analysis was performed using the JACoP plugin (Just Another Co-localization Plug-in; [40Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy J Microsc 2006; 224: 213-32.]) and the ImageJ software (http://rbs.info.nih.gov/ij/). The Mander´s overlap coefficient (OC), which expresses the correlation of pixel intensities in both analyzed channels was used for presentation of results. This coefficient ranges from 0 to 1, where 0 indicates no overlap and 1 reflects 100% colocalization between both channels of the analyzed image [41Manders E, Stap J, Brakenhoff G, van Driel R, Aten J. Dynamivs of three dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy J Cell Sci 1992; 103: 857-62.].
siRNA Induced HuR Knockdown
To knockdown endogenous expression of HuR protein in Huh7 cells, we used the pSIREN-RetroQ vector (BD Biosciences). The oligonucleotide sequences used for silencing the expression of HuR were predicted using a bioinformatic tool (http://bioinfo.clontech.com/rnaidesigner):
5´-gatccgaggcaattaccagtttcattcaagagatgaaactggtaattgcctctttttt acgcgtg 3’ (sense) and
5´-aattcacgcgtaaaaaagaggcaattaccagtttcatctcttgaatgaaactggtaa ttgcctcg-3’ (antisense), where the underlined nucleotides denote the HuR shRNA sequence targeting HuR mRNA (nucleotides 484 to 492 in HuR cDNA).
To construct plasmids used for RNAi the oligonucleotides were first allowed to anneal in vitro. This was performed by ressuspending in TE buffer followed by heating to 95ºC for 30 sec and two sequential incubations at 72º C and 37º C, both for 2 min. Finally the oligonucleotides were allowed to cool at room temperature to generate double stranded DNA (dsDNA). The obtained dsDNA was cloned into the EcoRI/BamHI digested pSIREN-RetroQ vector thus generating the pSIREN-RetroQ/HuR. To construct the control vector pSIREN-RetroQ/Luc, the oligonucleotides provided by the manufacturer for luciferase gene silencing were introduced into an identical vector according to the corresponding instructions.
For transfection, the FuGENE6 transfection reagent (Roche) was used according to the specifications of the manufacturer. First 1 µg plasmid pSVL(D3) was used to transfect Huh7 cells. After 48 hours incubation, cells were transfected again with plasmid pSIREN-RetroQ/HuR or the negative control (pSIREN-RetroQ/Luc). 24 hours post-transfection, puromycin was added to the medium (2µg/mL) to select for pSIREN-RetroQ plasmids transfected cells. Following additional 48 hours incubation, cells were harvested for subsequent western blot analysis. Western blots were performed as described before using the following primary antibodies: rabbit polyclonal anti-HDAg antibody (B3; [37Saldanha J, Homer E, Goldin P, Thomas HC, Monjardino J. Cloning and expression of an immunodominant region of the hepatitis delta antigen J Gen Virol 1990; 71: 471-5.]), mouse anti-His-tag monoclonal antibody (Sigma-Aldrich), rabbit polyclonal anti-clathrin antibody (Santa Cruz Biotechnology), and a mouse monoclonal antibody against HuR (Santa Cruz Biotechnology). After incubation with appropriate anti-mouse or anti-rabbit secondary antibodies conjugated with horseradish peroxidase (GE Healthcare) blots were developed using an ECL system (GE Healthcare).
RESULTS
Identification of HuR Protein as a Binding Partner of S-HDAg in a Yeast Two-Hybrid Screening
To identify proteins capable of interacting with S-HDAg, this protein was fused to the C terminus of the DNA binding domain of GAL4 (GAL4BD) and used as bait in a yeast two hybrid screen of a human liver cDNA library containing approximately 1 x108 clones. A total of 112 positive clones were obtained. After sequence analysis and database search, one of the positive clones was found to bear a plasmid containing a cDNA fragment encoding about 96% of the entire open reading frame (ORF) of the ELAV-like 1 (ELAVL1) protein, in frame with the GAL4 activation domain (Fig. 1A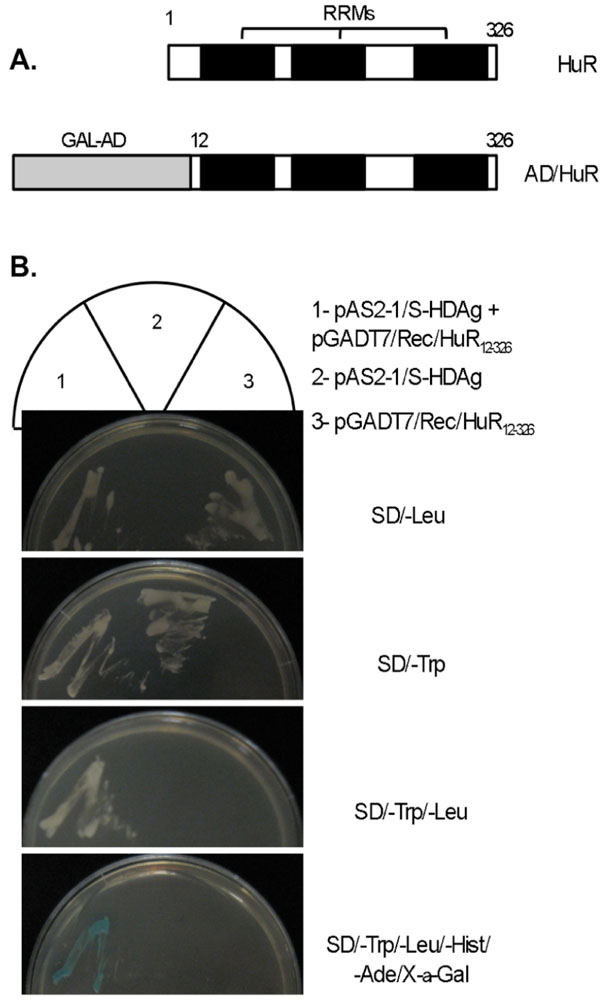 ).
).
Human ELAVL1 (GenBank NP_001419), also known as HuR (Hu antigen R), is a 326 amino acid protein and has been described as a RNA binding protein, comprising three RNA binding domains (RBDs), also known as RNA recognition motifs (RRMs). The sequence identified in the yeast two- hybrid screening comprises amino acids 12 to 326 of the full length HuR (Fig. 1A ), including the three RRMs.
), including the three RRMs.
The HuR protein was previously reported to participate in post-transcriptional regulation processes and has also been proposed to be an important player in the replication cycle of several viruses, including HIV-1, HCV, and HVS [30Lemay J, Mandou-Peindara P, Bader T, et al. HuR interacts with human immunodeficiency virus type I reverse transcriptase, and modulates reverse transcription in infected cells Retrovirology 2008; 5: 47., 31Spangberg K, Wiklund L, Schwartz S. HuR, a protein implicated in oncogene and growth factor mRNA decay, binds to the 3’ ends of hepatitis C virus RNA of both polarities Virology 2000; 274: 378-90., 42Cook HL, Mischo HE, Steitz JA. The Herpesvirus saimiri small nuclear RNAs recruit AU-rich element-containing mRNA levels in virally transformed T cells Mol Cell Biol 2004; 24: 4522-33.]. Thus, it is possible that this protein might also involved in HDV replication. To investigate this hypothesis we first decided to confirm the results obtained in the yeast two-hybrid screening. Yeast AH109 cells were cotransformed with the bait (pAS2-1/S-HDAg) and prey plasmids isolated from the positive yeast clone (pGADT7/Rec/HuR12-326), and the interaction between the two proteins was tested using three reporter genes (ADE2, HIS3, and MEL1). Yeast AH109 cells transformed with pAS2-1/S-HDAg or pGADT7-Rec/HuR12-326 alone were used as controls in these experiments. In contrast with the negative controls, yeast cells expressing both BD/S-HDAg and AD/HuR12-326 fusion proteins could effectively grow in minimal medium lacking tryptophan, leucine, histidine, and adenine (SD/-Leu/-Trp/-Hist/-Ade), and displayed strong α-galactosidase activity (Fig. 1B ). These results allowed confirming the interaction of S-HDAg and HuR in the yeast two-hybrid system.
). These results allowed confirming the interaction of S-HDAg and HuR in the yeast two-hybrid system.
HuR and S-HDAg Interact In Vitro and In Vivo in Huh7 Cells
After establishing that S-HDAg and HuR interact in the yeast two-hybrid system, we decided to investigate whether the two proteins also interact in human liver cells. We first tested this interaction in vitro using a blot overlay assay. The vector pGEX-6P-2/S-HDAg was used to produce GST/S-HDAg fusion protein in E. coli cells. GST/S-HDAg protein was purified from bacterial extracts using glutathione sepharose columns and the GST tag was cleaved with PreScission Protease to obtain untagged S-HDAg. Purification of recombinant untagged S-HDAg was evaluated by western blot, and protein concentration was determined using a Bradford assay (data not shown). Expression of His6-HuR was induced in E. coli cells transformed with plasmid pET28c/HuR. Increasing amounts of the obtained protein samples were loaded onto different wells of a 12% SDS-PAGE gel. After separation proteins were transferred to a nitrocellulose membrane which was subsequently overlaid with 40 µg of recombinant S-HDAg. Detection of bound S-HDAg was performed using a specific polyclonal antibody. The obtained results are displayed in Fig. (2A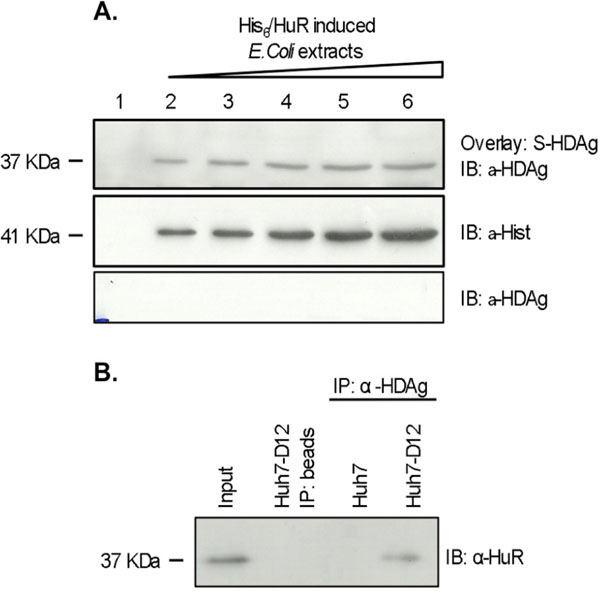 ) showing that increasing amounts of the His6-HuR protein result in a proportional increase in the amounts of bound S-HDAg, suggesting a specific interaction between the two proteins.
) showing that increasing amounts of the His6-HuR protein result in a proportional increase in the amounts of bound S-HDAg, suggesting a specific interaction between the two proteins.
The in vivo interaction of HuR with S-HDAg was further investigated using a coimmunoprecipitation assay and Huh7-D12 cells. This cell line was previously reported to express constitutively both forms of the delta antigen and virus RNA [35Cheng D, Yang A, Thomas H, Monjardino J. Characterization of stable hepatitis delta-expressing cell lines: Effect of HDAg on cell growth Prog Clin Biol Res 1993; 382: 149-53., 38Cunha C, Monjardino J, Chang D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus in the nucleus of human cells RNA 1998; 4: 680-93.]. Since HuR is ubiquitously expressed in proliferating cells [43Ma WJ, Cheng S, Campbell C, Wrigth A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein J Biol Chem 1996; 271: 8144-51.], the eventual interaction of the two proteins should be detectable in this system. Huh7-D12 protein extracts were prepared and incubated with a rabbit polyclonal antibody against HDAg. Immunoprecipitated protein samples were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and immunobloted with an anti-HuR antibody. Similar procedures in the absence of anti-HDAg antibodies or using Huh7 cells protein extracts were performed in these experiments as negative controls. As shown in Fig. (2B ), in Huh7-D12 cells protein extracts HuR was coprecipitated together with the S-HDAg being consistent with an in vivo association of the two proteins in human liver cells.
), in Huh7-D12 cells protein extracts HuR was coprecipitated together with the S-HDAg being consistent with an in vivo association of the two proteins in human liver cells.
HuR Colocalizes with HDV Antigens and RNA in Human Hepatoma Cells
It has been previously shown that the HuR protein is involved in post-transcriptional modifications of RNAs and may modulate the replication of HIV-1, HCV, HSV, and HPV [30Lemay J, Mandou-Peindara P, Bader T, et al. HuR interacts with human immunodeficiency virus type I reverse transcriptase, and modulates reverse transcription in infected cells Retrovirology 2008; 5: 47.-33Rivas-Aravena A, Ramdohr P, Vallejos M, et al. The ELAV-like protein HuR exerts translational control of viral internal ribosome entry sites Virology 2009; 392: 178-85.]. Since S-HDAg and HuR interact both in vitro and in vivo, it may be plausible that HuR is also involved in the control of some stages of the HDV replication cycle. To test this possibility our first approach was designed in order to investigate an eventual colocalization of HuR with the virus RNA and antigens inside the cell. This was performed by immunofluorescence and in situ hybridization followed by confocal microscopy and image analysis of Huh7-D12 cells, which constitutively express HDV RNPs [35Cheng D, Yang A, Thomas H, Monjardino J. Characterization of stable hepatitis delta-expressing cell lines: Effect of HDAg on cell growth Prog Clin Biol Res 1993; 382: 149-53., 38Cunha C, Monjardino J, Chang D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus in the nucleus of human cells RNA 1998; 4: 680-93.].
In accordance with previous findings [38Cunha C, Monjardino J, Chang D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus in the nucleus of human cells RNA 1998; 4: 680-93.] the delta antigens were found to be localized in the nucleus of Huh7-D12 cells, with a preferential accumulation in foci (Fig. 3A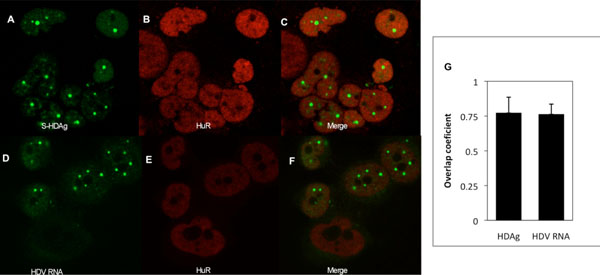 ). The HuR protein also showed a nuclear localization pattern and seemed to colocalize with the HDAgs (Fig. 3B
). The HuR protein also showed a nuclear localization pattern and seemed to colocalize with the HDAgs (Fig. 3B , C
, C ). The extent of colocalization of S-HDAg and HuR was analyzed quantitatively by calculating the Mander´s overlap coefficient (OC). This coefficient gives an estimate of intensity-based correlation of green (HDAg) and red (HuR) signals. The average OC value (n=10) for HuR and S-HDAg is 0.772 (std. deviation= 0,113; n=10) indicating a significant colocalization. The results obtained are consistent with the hypothesis that HuR and HDAg physically interact in Huh7-D12 cells.
). The extent of colocalization of S-HDAg and HuR was analyzed quantitatively by calculating the Mander´s overlap coefficient (OC). This coefficient gives an estimate of intensity-based correlation of green (HDAg) and red (HuR) signals. The average OC value (n=10) for HuR and S-HDAg is 0.772 (std. deviation= 0,113; n=10) indicating a significant colocalization. The results obtained are consistent with the hypothesis that HuR and HDAg physically interact in Huh7-D12 cells.
Previous reports showed that HDAgs and HDV RNA colocalize in the nucleus of human liver cells [38Cunha C, Monjardino J, Chang D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus in the nucleus of human cells RNA 1998; 4: 680-93.]. Since HuR is a RNA binding protein we decided to investigate if the distribution pattern of the protein in relation to the virus RNA. To do this, a combined in situ hybridization and immunofluorescence approach was utilized. A digoxigenin-labelled probe was used to hybridize with HDV RNAs and the HuR protein was detected by immunofluorescence, as above described. Typical examples of the obtained results can be observed in Fig. (3D , F
, F ). The average OC value obtained (0.762; std. deviation = 0.074; n=10), is very similar with the one determined for HuR/S-HDAg colocalization. Taken together, our results indicate that the HuR protein interacts with S-HDAg in human liver cells, and additionally suggest that it may also interact with the virus RNA.
). The average OC value obtained (0.762; std. deviation = 0.074; n=10), is very similar with the one determined for HuR/S-HDAg colocalization. Taken together, our results indicate that the HuR protein interacts with S-HDAg in human liver cells, and additionally suggest that it may also interact with the virus RNA.
HuR Protein is Overexpressed in HDV Expressing Cells
It is known that viruses subvert the cellular metabolism namely by recruiting host factors to locals of replication in the nucleus and by altering gene expression in infected cells. Accordingly, it has been previously reported that in human hepatoma cells, synthesis of HDV RNA and antigens is accompanied by changes in expression levels of several host proteins [21Mota S, Mendes M, Penque D, Coelho AV, Cunha C. Changes in the proteome of Huh7 cells induced by transient expression of hepatitis D virus RNA and antigens J Proteomics 2008; 71: 71-9., 22Mota S, Mendes M, Freitas N, Penque D, Coelho AV, Cunha C. Proteome analysis of a human liver carcinoma cell line stably expressing hepatitis delta vírus ribonucleoproteins J Proteomics 2009; 72: 616-27.]
In an attempt to investigate if the synthesis of HDV RNPs also induces alterations in the expression levels of the HuR protein we decided to compare the relative amounts of the protein in Huh7 and Huh7-D12 cells. Protein extracts from both cell lines were prepared, separated by SDS-PAGE, and analyzed by western blot using an antibody to detect the HuR protein. An antibody recognizing the housekeeping protein clathrin was used as an internal control in these experiments. The blot images were digitalized and band intensities were calculated by densitometry using the ImageJ program. The obtained results showed that the expression levels of HuR were increased 1.7-fold in Huh7-D12 cells when compared with Huh7 cells (Fig. 4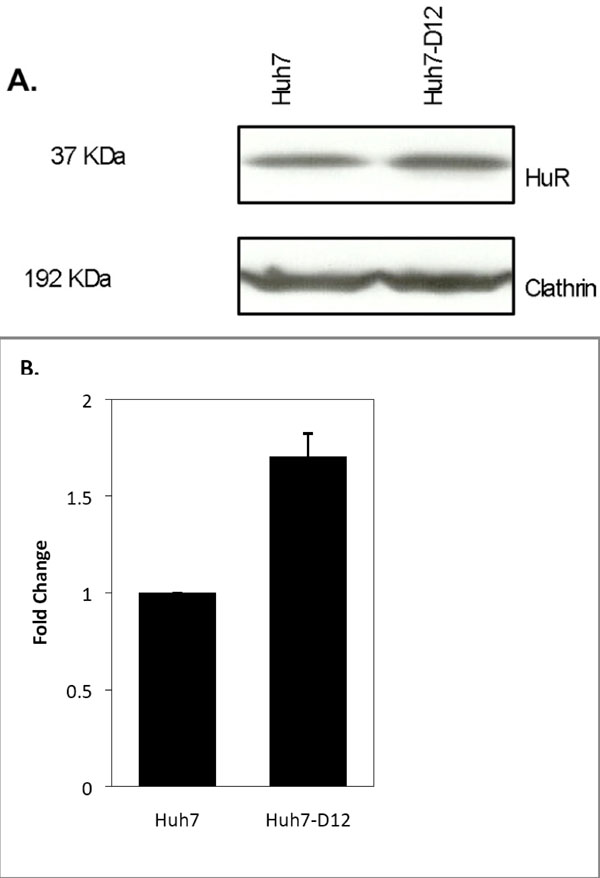 ). Considering that only 10-15% of Huh-D12 cells are expressing HDV RNPs at a given moment [38Cunha C, Monjardino J, Chang D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus in the nucleus of human cells RNA 1998; 4: 680-93.] this finding may be indicative of a possible involvement of the HuR protein in the virus replication cycle.
). Considering that only 10-15% of Huh-D12 cells are expressing HDV RNPs at a given moment [38Cunha C, Monjardino J, Chang D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus in the nucleus of human cells RNA 1998; 4: 680-93.] this finding may be indicative of a possible involvement of the HuR protein in the virus replication cycle.
Silencing of the HuR Protein Results in Decreased Expression of HDAgs
The HuR protein was previously shown to play an important role in mRNA stabilization and control of translation (reviewed in [25Brennan CM, Steitz JA. HuR and mRNA stability Cell Mol Life Sci 2001; 58: 266-77.]). Since we found that HuR binds in vivo to S-HDAg, both proteins colocalize in Huh7 cells, and HuR is overexpressed during synthesis of HDV RNPs, we decided to investigate the effect of HuR knockdown on HDAg synthesis.
To do this we utilized a small hairpin RNA mediated knockdown technology based on a pSIREN-RetroQ vector coding for a HuR-specific short hairpin RNA (pSIREN-RetroQ/HuR). Huh7 cells were first transfected with plasmid pSVL(D3) in order to produce HDV RNA and antigens, and 48 hours later transfected again with the pSIREN-RetroQ-HuR plasmid. Puromycin was added to the medium in order to select the pSIREN-RetroQ/HuR transfected cells, and protein extracts were prepared. A pSIREN-RetroQ vector, targeting the luciferase mRNA (pSIREN-RetroQ/Luc) was used in parallel transfections and used as negative control in these experiments. The efficiency of reduction of cellular HuR protein levels by the shRNAs coded by the pSIREN-RetroQ/Hur vector was monitored by western blot. As shown in Fig. (5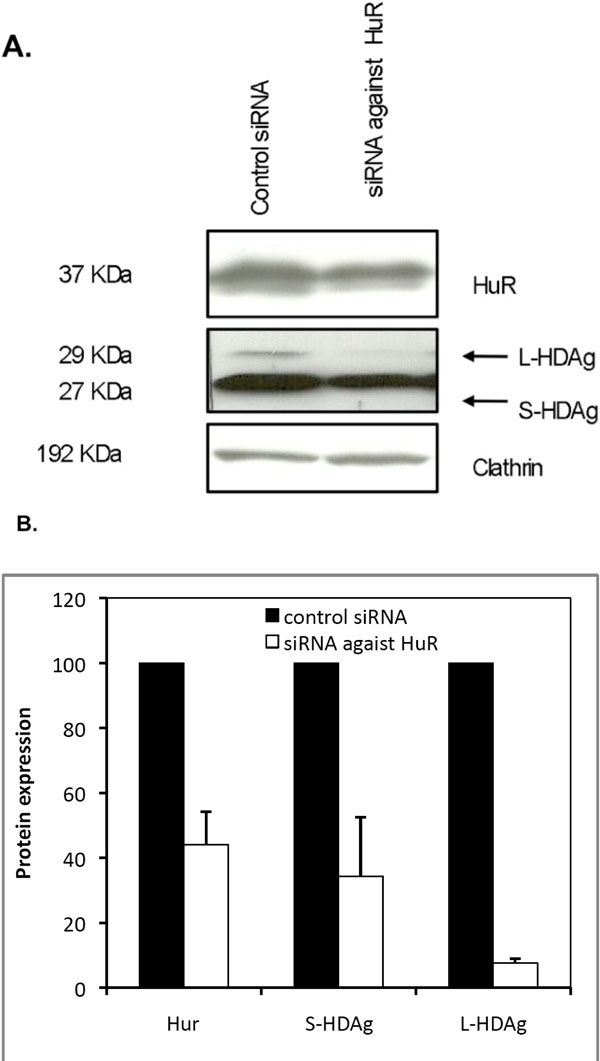 ), the plasmid-derived expression of shRNA against HuR mRNA, reduced the expression of the HuR protein by 55% when compared with control pSIREN-RetroQ/Luc transfected cells.
), the plasmid-derived expression of shRNA against HuR mRNA, reduced the expression of the HuR protein by 55% when compared with control pSIREN-RetroQ/Luc transfected cells.
We next investigated the effect of HuR silencing in the synthesis of delta antigens. This was performed by analyzing the relative expression of HDAgs by western blot of protein samples obtained from the same knockdown experiment. The results are displayed in Fig. (5 ) and show that knockdown of HuR resulted in a decrease of 65% and 93%, in S-HDAg and L-HDAg expression levels, respectively. These results indicate that HDAg expression is inhibited when HuR is silenced in Huh7 cells, suggesting that this protein may play an important role in HDV replication.
) and show that knockdown of HuR resulted in a decrease of 65% and 93%, in S-HDAg and L-HDAg expression levels, respectively. These results indicate that HDAg expression is inhibited when HuR is silenced in Huh7 cells, suggesting that this protein may play an important role in HDV replication.
DISCUSSION
Hepatitis delta virus is the smallest human pathogen identified so far being highly dependent on host factors for replication and transcription. The HDV RNA genome contains a single ORF from which two forms, small and large, of a unique protein are derived as a consequence of an editing mechanism. Despite its simplicity a limited number of information is still available concerning the cellular proteins that interact with HDV RNA or antigens to modulate replication and pathogenesis. Among them are RNA polymerases I, II, and III, the small form of adenosine deaminase (ADAR 1), hnRNP-L, ASF/SF2, p54 (nrb), GADPH, eEF1A1, NELF, PSF, PKR, nucleolin, and DIPA (reviewed in [12Greco-Stewart VS, Pelchat M. Interaction of Host Cellular proteins with components of the Hepatitis Delta virus Viruses 2010; 2: 189-212.]). Recently, using a combined proteomic-RNAi screen of potential host factors involved in HDV replication it was possible to identify over 100 proteins capable of binding to FLAG-S-HDAg [23Cao D, Haussecker D, Huang Y, Kay MA. Combined proteomic-RNAi screen for host factors involved in human hepatitis delta virus replication RNA 2009; 15: 1971-9.].
In this work, we used a yeast two-hybrid approach to identify human liver proteins capable of interacting with S-HDAg. We initially obtained 112 positive clones of which 31 were found to contain DNA sequences encoding for known proteins in a database (Genbank). One of these sequences was found to code for the HuR protein, a member of the ELAV family of RNA binding proteins. Subsequently, this sequence was subcloned in a prokaryotic expression vector (pET28c) and the corresponding protein was produced in E. coli as a fusion with His-tag. HuR is a nucleocyto-plasmic shuttling protein previously shown to be involved in mRNA stabilization. It is well established that HuR, which is ubiquously expressed in proliferating cells, and selectively binds AU-rich elements (AREs) in the 3’ untranslated region of mRNAs. Binding of HuR to mRNAs is believed to antagonize the mRNA degradation pathway signaled by AREs. The Herpesvirus saimiri small nuclear RNAs contain an ARE sequence which was found to be able to recruit HuR [42Cook HL, Mischo HE, Steitz JA. The Herpesvirus saimiri small nuclear RNAs recruit AU-rich element-containing mRNA levels in virally transformed T cells Mol Cell Biol 2004; 24: 4522-33.]. Furthermore, HuR was reported to modulate translation of HIV-1 and HCV IRES elements acting as a repressor or activator, respectively [33Rivas-Aravena A, Ramdohr P, Vallejos M, et al. The ELAV-like protein HuR exerts translational control of viral internal ribosome entry sites Virology 2009; 392: 178-85.]. This regulation does not require direct binding of HuR to IRES.
This prompted us to investigate a possible involvement of HuR in HDV replication. Our first approach was directed to confirm the detected HuR-S-HDAg interaction observed in the yeast two-hybrid screening. Initially, this was performed using a blot overlay assay with purified S-HDAg and increasing amounts of HuR protein present in E. coli cell extracts. The results obtained with this assay indicated that HuR and S-HDAg specifically interact in vitro. Finally, we decided to investigate if the two proteins interact in vivo in human liver cells. A coimmunoprecipitation assay using a specific polyclonal antibody against HDAg was performed allowing confirming the previously observed interactions.
It is known that viruses subvert the nuclear organization and can recruit components of the host cell machinery to sites of replication and transcription (for a review see [44Ahlquist P, Noueiry AO, Lee W-M, Kushner DB, Dye BT. Host Factors in Positive-Strand RNA Virus Genome Replication J Virol 2003; 77: 8181-6.]). In the particular case of HDV, a previous work showed that both delta antigens and RNA are mainly localized in the nucleus of infected cells displaying, in most cases, a diffuse nucleoplasmic distribution with additional accumulation in foci [38Cunha C, Monjardino J, Chang D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus in the nucleus of human cells RNA 1998; 4: 680-93.]. As shown by the authors, these foci although containing virus RNA and antigens, do not represent preferential sites of HDV replication and transcription. However, if HuR is involved in HDV replication one would expect to observe a colocalization of this protein with HDV components in the nucleus. This hypothesis was tested by immunofluorescence and in situ hybridization in Huh7-D12 cells, and HDV cDNA stably transfected human hepatoma cell line which constitutively expresses both virus RNA and antigens [35Cheng D, Yang A, Thomas H, Monjardino J. Characterization of stable hepatitis delta-expressing cell lines: Effect of HDAg on cell growth Prog Clin Biol Res 1993; 382: 149-53., 38Cunha C, Monjardino J, Chang D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus in the nucleus of human cells RNA 1998; 4: 680-93.]. After confocal microscopy followed by image analysis to calculate the Mander’s overlap coefficient, it was found that over 75% of endogenous HuR protein colocalizes with both virus RNA and antigens indicating that HuR may play a role in the HDV life cycle.
Previous proteomic analysis of Huh7 cells stably expressing HDV RNPs or transiently expressing each of the HDV components separately, allowed identifying several host proteins displaying altered expression patterns [21Mota S, Mendes M, Penque D, Coelho AV, Cunha C. Changes in the proteome of Huh7 cells induced by transient expression of hepatitis D virus RNA and antigens J Proteomics 2008; 71: 71-9., 22Mota S, Mendes M, Freitas N, Penque D, Coelho AV, Cunha C. Proteome analysis of a human liver carcinoma cell line stably expressing hepatitis delta vírus ribonucleoproteins J Proteomics 2009; 72: 616-27.]. Although the authors did not address possible interactions with host proteins, more recently it was reported that two of the differentially expressed proteins in S-HDAg transfected Huh7 cells, hnRNP D and ZNF326, interact with S-HDAg [23Cao D, Haussecker D, Huang Y, Kay MA. Combined proteomic-RNAi screen for host factors involved in human hepatitis delta virus replication RNA 2009; 15: 1971-9.]. In an attempt to investigate if the HuR protein is also differentially expressed in cells expressing HDV components, we compared the relative amounts of this protein in Huh7 and Huh7-D12 cells. Western blots were performed in order to detect HuR followed by determination of band intensity. The results were normalized using an antibody against the housekeeping protein clathrin. We found that HuR is 1.7 times more abundant in HDV expressing Huh7-D12 cells when compared to Huh7 cells. As reported before, only a limited amount, 10%-15% of Huh7-D12 cells express HDV RNPs at a given moment in time [38Cunha C, Monjardino J, Chang D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus in the nucleus of human cells RNA 1998; 4: 680-93.]. Accordingly, the relative expression values of HuR in this cell line may be indicative of a significant upregulation in the presence of HDV.
Silencing of HuR with a specific siRNA resulted in marked decrease of both S-HDAg and L-HDAg expression suggesting a possible role in HDV replication. However, the mechanism(s) by which HuR influences the expression of delta antigens remain to be elucidated. Since HDV mRNAs are capped and polyadenylated [45Gudima S, Dingle K, Wu TT, Moraleda G, Taylor J. Characterization of the 5’ ends for polyadenilated RNAs synthesized during replication of hepatitis delta virus J Virol 1999; 73: 6533-9.] it is unlikely that HuR participates in their stabilization by binding to AREs. It is, thus, possible to speculate that the effect of HuR silencing on HDAg expression may be due to an impaired stabilization of cellular mRNAs with consequent decrease in production of host proteins necessary for HDV replication. This may be supported by reports showing that HuR stabilizes ARE-containing mRNAs [46Brennan CM, Gallouzi I-E, Steitz JA. Protein ligands to HuR modulate its interaction with targets mRNAs in vivo J Cell Biol 2000; 151: 1-14.] namely in response to stress induced by heat-shock, UV light and infection [47Wang W, Furneaux H, Cheng H, et al. HuR regulates p21 mRNA stabilization by UV light Mol Cell Biol 2000; 20: 760-9.]. Alternatively, HuR could contribute to the stabilizion of genomic and/or antigenomic HDV RNAs during replication and transcription.
The mechanisms by which HuR promotes mRNA stability were previously addressed allowing the identification of four protein ligands in coimmunoprecipitation assays: SETα, SETβ, pp32, and APRIL [46Brennan CM, Gallouzi I-E, Steitz JA. Protein ligands to HuR modulate its interaction with targets mRNAs in vivo J Cell Biol 2000; 151: 1-14.]. Binding of these ligands to HuR is thought to increase HuR affinity for mRNAs or modulate its intracellular localization. Since HuR function is altered depending on the bound protein ligand it is temptative to suggest that the interaction between S-HDAg and HuR may also influence the activity of the protein(s) modulating HDV replication. In accordance with this view, HuR was also found to interact with the HIV-1 reverse transcriptase, modulating the activity of the enzyme, in a mechanism independent of binding of HuR to the virus RNA [30Lemay J, Mandou-Peindara P, Bader T, et al. HuR interacts with human immunodeficiency virus type I reverse transcriptase, and modulates reverse transcription in infected cells Retrovirology 2008; 5: 47.]. Several functions have been assigned to the S-HDAg in virus replication cycle: stabilization of the genome and antigenome [23Cao D, Haussecker D, Huang Y, Kay MA. Combined proteomic-RNAi screen for host factors involved in human hepatitis delta virus replication RNA 2009; 15: 1971-9., 48Lazinski DW, Taylor JM. Intracellular cleavage and ligation of hepatitis delta virus genomic RNA: Regulation of ribozyme activity by cis-acting sequences and host factors J Virol 1995; 69: 1190-200.], protection of HDV RNAs against editing [49Cheng Q, Jayan GC, Casey JL. Differential inhibition of RNA editing in hepatitis delta virus genotype III by the short and long forms of hepatitis delta antigen J Virol 2003; 77: 7786-95.], transport of the virus genome to the nucleus [50Xia YP, Yeh CT, Ou JH, Lai MMC. Characterization of nuclear targeting signal of hepatitis delta antigen: nuclear transport as a protein complex J Virol 1992; 66: 914-21.], acceleration of ribozyme activities [51Huang ZS, Wu HN. Identification and characterization of the RNA chaperone activity of hepatitis delta antigen peptides J Biol Chem 1998; 273: 26455-61., 52Jeng KS, Su PY, Lai MM. Hepatitis delta antigens enhance the ribozyme activities of hepatitis delta virus RNA in vivo J Virol 1996; 70: 4205-9.], and improvement of processivity during RNA-directed RNA transcription [10Yamaguchi Y, Filipovska J, Yano K, et al. Stimulation of RNA polymerase II elongation by hepatitis delta antigen Science 2001; 293: 124-7.]. Whether binding of HuR affects, positively or negatively, any of the S-HDAg functions remains to be clarified and clearly needs to be addressed in the future.
CONCLUSION
In conclusion, we report that HuR and S-HDAg interact both in vitro and in vivo and colocalize in human liver cells. Furthermore, HuR silencing with specific siRNAs leads to a decrease in both S-HDAg and L-HDAg expression. The obtained results suggest that HuR may play an important role in HDV replication.
ACKNOWLEDGMENTS
We are grateful to Dr. John Taylor (Fox Chase Cancer Center, USA) for the generous gift of plasmid pSVL(D3) used in this work. We acknowledge Dr. José Rino (Institute of Molecular Medicine, Lisbon) for help with the confocal microscope. This work was supported by Fundação para a Ciência e Tecnologia (PTDC/SAU-MII/098314/2008). AC is a recipient of a FCT PhD grant.







