- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
HTS-Compatible β-Lactamase Transcriptional Reporter Gene Assay for Interrogating the Heat Shock Response Pathway
Michael K Hancock1, Menghang Xia2, Elizabeth S Frey1, Srilatha Sakamuru2, Kun Bi*, 1
Abstract
Moderate environmental and physiological stressors are known to initiate protective heat shock response (HSR) leading to cell survival. HSR is largely mediated by the activation of heat shock factor (HSF), resulting in increased heat shock protein expression. Dysregulation of the HSR signaling has been associated with various diseases including cancer, inflammation and neurodegenerative disorders. Compounds that can modulate HSR have been pursued for the treatment of these diseases. To facilitate the discovery of HSR modulators, we developed a high-throughput amenable betalactamase transcriptional reporter gene assay for monitoring the function of HSF. HeLa cells were engineered to express the beta-lactamase reporter under the control of HSF response elements (HSE) present in the HSP70 gene promoter. The HSE-beta lactamase (HSE-bla) reporter gene assay was validated by using HSF-specific siRNAs and known small molecule modulators. Taking the advantage of fluorescence resonance energy transfer (FRET)-based cell permeable betalactamase substrate, this assay can be miniaturized into 1536-well format. Our results demonstrate that the assay is robust and can be applied to high-throughput screening (HTS) for modulators of HSR.
Article Information
Identifiers and Pagination:
Year: 2009Volume: 3
First Page: 1
Last Page: 6
Publisher Id: CCGTM-3-1
DOI: 10.2174/1875397300903010001
Article History:
Received Date: 09/11/2008Revision Received Date: 10/12/2008
Acceptance Date: 14/12/2008
Electronic publication date: 05/2/2009
Collection year: 2009
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.5/), which permits unrestrictive use, distribution, and reproduction in any medium, provided the original work is properly cited.
* Address correspondence to this author at the Invitrogen Corporation, Discovery Assays and Services, 501 Charmany Drive, Madison, WI 53719, USA; E-mail: kun.bi@invitrogen.com
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 09-11-2008 |
Original Manuscript | HTS-Compatible β-Lactamase Transcriptional Reporter Gene Assay for Interrogating the Heat Shock Response Pathway | |
INTRODUCTION
Environmental and physiological stressors such as elevated temperature, chemical toxicants and oxidative stress are known to induce heat shock response (HSR) leading to the increased expression of molecular chaperones such as heat shock proteins (HSPs) and other cytoprotective proteins [1Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation J Biol Chem 2005; 280: 33097-100.]. HSR and the induction of HSPs are mediated by a family of transcription factors called heat shock factors (HSFs), which bind to the heat shock elements (HSEs) present in the promoter regions of HSP genes [1Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation J Biol Chem 2005; 280: 33097-100.]. Among the three human HSFs, HSF-1, -2, and -4, HSF1 is the best characterized and essential for HSR [2Voellmy R. Feedback regulation of the heat shock response Handb Exp Pharmacol 2006; 43-68.]. Under normal conditions HSF1 exists as an inert monomer held in a repressed state, presumably by interaction with HSP90 and HSP70 [1Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation J Biol Chem 2005; 280: 33097-100., 3Powers MV, Workman P. Inhibitors of the heat shock response: biology and pharmacology FEBS Lett 2007; 581: 3758-69.]. Upon exposure to stress, the rising level of unfolded proteins results in the dissociation and derepression of HSF1. HSF1 then trimerizes, undergoes post-translational modifications, and accumulates in the nucleus where it activates HSP gene expression (Fig. 1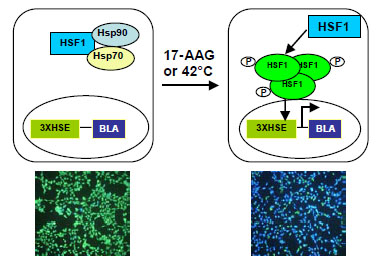 ) [3Powers MV, Workman P. Inhibitors of the heat shock response: biology and pharmacology FEBS Lett 2007; 581: 3758-69.].
) [3Powers MV, Workman P. Inhibitors of the heat shock response: biology and pharmacology FEBS Lett 2007; 581: 3758-69.].
 |
Fig (1) Diagram of the cell line design for HSR. |
Aberrations in HSR have been associated with various human diseases, such as neurodegenerative disorder, inflammation and cancer [4Kim HJ, Hwang NR, Lee KJ. Heat shock responses for understanding diseases of protein denaturation Mol Cell 2007; 23: 123-31.]. Small molecule modulators of HSR including those directly targeting HSP chaperones have been pursued for the treatment of these diseases [4Kim HJ, Hwang NR, Lee KJ. Heat shock responses for understanding diseases of protein denaturation Mol Cell 2007; 23: 123-31.]. For example, HSP90 inhibitor 17-allylamino-17-demethoxygel-danamycin (17-AAG) is currently in phase II clinical trial in patients with various cancers including melanoma and breast cancer [4Kim HJ, Hwang NR, Lee KJ. Heat shock responses for understanding diseases of protein denaturation Mol Cell 2007; 23: 123-31.]. Importantly, inhibition of HSP90 by 17-AAG also derepressses HSF1 and induces HSR [5Bagatell R, Paine-Murrieta GD, Taylor CW, et al. Induction of a heat shock factor 1-dependent stress response alters the cytotoxic activity of hsp90-binding agents Clin Cancer Res 2000; 6: 3312-18.]. Besides inhibitors of HSP90, proteasome inhibitors, amino acid analogues, and molecules such as arachidonic acid and prostaglandins that induce inflammatory responses have been reported to induce HSR [1Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation J Biol Chem 2005; 280: 33097-100., 4Kim HJ, Hwang NR, Lee KJ. Heat shock responses for understanding diseases of protein denaturation Mol Cell 2007; 23: 123-31.]. Such inducers of HSR may prove beneficial to counteracting the protein misfolding and aggregation associated with various neurodegenerative disorders.
On the other hand, since tumor cells often express high levels of HSPs to survive the stressful environment in solid tumors, inhibitors of the HSR could be beneficial for cancer patients. Several inhibitors of the HSR have been reported. Quercetin (3,3’,4’,5,7-pentahydroxyflavone) is a natural compound that inhibits HSR in many cell types by down-regulating HSF1 activity [6Nagai N, Nakai A, Nagata K. Quercetin suppresses heat shock response by down regulation of HSF1 Biochem Biophys Res Commun 1995; 208: 1099-05.]. Benzylidene lactam KNK437 is reported to be a more selective compound that inhibits the expression of certain HSP family members [7Yokota S, Kitahara M, Nagata K. Benzylidene lactam compound, KNK437, a novel inhibitor of acquisition of thermotolerance and heat shock protein induction in human colon carcinoma cells Cancer Res 2000; 60: 2942-48.]. Both quercetin and KNK437 inhibit cancer proliferation and render the cells more sensitive to hyperthermia and chemotherapy [6Nagai N, Nakai A, Nagata K. Quercetin suppresses heat shock response by down regulation of HSF1 Biochem Biophys Res Commun 1995; 208: 1099-05., 7Yokota S, Kitahara M, Nagata K. Benzylidene lactam compound, KNK437, a novel inhibitor of acquisition of thermotolerance and heat shock protein induction in human colon carcinoma cells Cancer Res 2000; 60: 2942-48.]. Other reported inhibitors of HSR include triptolide, NZ28 and emunin, each with different mechanisms of action [4Kim HJ, Hwang NR, Lee KJ. Heat shock responses for understanding diseases of protein denaturation Mol Cell 2007; 23: 123-31.]. In order to move forward for clinical development a significant effort needs to be invested to improve the potency and selectivity of these compounds.
The lack of specific HSR modulators is partially due to a lack of robust cell-based assay tools for high-throughput screening. Here, we describe the development of a reporter gene assay for HSR applicable to high-throughput screening.HeLa cells were engineered to express the beta-lactamase reporter gene [8Zlokarnik G, Negulescu PA, Knapp TE, et al. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter Science 1998; 279: 84-8.] under the control of HSF response elements present in the HSP70 gene promoter. The HSE-bla reporter cell line was validated using HSF1-specific siRNAs and known small molecule modulators, and the assay was miniaturized into 1536-well format. Our results demonstrate that the assay is robust and can be applied to high-throughput screening for modulators of HSR.
PRINCIPLE OF ASSAY DESIGN
HeLa cells were transduced with lentivirus to express a beta-lactamase reporter gene under the control of three copies of HSE sequence (CTGGAATATTCCCGACCT GGCAG) present in the HSP70 gene promoter (Fig. 1 ). Cells expressing beta-lactamase after 42°C heat shock were isolated using fluorescence-activated cell sorting (FACS). In its un-induced state HSF1 exists as an inert monomer that is presumably repressed by interactions with HSP90 and HSP70 (Fig. 1
). Cells expressing beta-lactamase after 42°C heat shock were isolated using fluorescence-activated cell sorting (FACS). In its un-induced state HSF1 exists as an inert monomer that is presumably repressed by interactions with HSP90 and HSP70 (Fig. 1 ) [1Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation J Biol Chem 2005; 280: 33097-100., 3Powers MV, Workman P. Inhibitors of the heat shock response: biology and pharmacology FEBS Lett 2007; 581: 3758-69.]. Heat shock or a known HSR inducer such as 17-AAG treatment leads to trimerization and phosphorylation of HSF1, which in turn binds to DNA response elements driving the expression of the downstream beta-lactamase reporter gene (Fig. 1
) [1Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation J Biol Chem 2005; 280: 33097-100., 3Powers MV, Workman P. Inhibitors of the heat shock response: biology and pharmacology FEBS Lett 2007; 581: 3758-69.]. Heat shock or a known HSR inducer such as 17-AAG treatment leads to trimerization and phosphorylation of HSF1, which in turn binds to DNA response elements driving the expression of the downstream beta-lactamase reporter gene (Fig. 1 ). Beta-lactamase activity can be detected by adding its substrate CCF4-AM directly into the cells [8Zlokarnik G, Negulescu PA, Knapp TE, et al. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter Science 1998; 279: 84-8., 9Wang J, Hancock MK, Dudek JM, Bi K. Cellular assays for high-throughput screening for modulators of Trk receptor tyrosine kinases Curr Chem Genomics 2008; 1: 27-33.]. In the absence of HSR inducer the pathway is inactive, therefore there is little beta-lactamase expression. When the cells are loaded with CCF4-AM and are excited at 406 nm, the substrate emits at around 530 nm (green). However, when cells are stimulated with 17-AAG the pathway is activated, leading to beta-lactamase reporter gene expression. The substrate is then cleaved by beta-lactamase, thereby disrupting the fluorescence resonance energy transfer (FRET) and resulting in emission at 460 nm (blue) following excitation at 406 nm.
). Beta-lactamase activity can be detected by adding its substrate CCF4-AM directly into the cells [8Zlokarnik G, Negulescu PA, Knapp TE, et al. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter Science 1998; 279: 84-8., 9Wang J, Hancock MK, Dudek JM, Bi K. Cellular assays for high-throughput screening for modulators of Trk receptor tyrosine kinases Curr Chem Genomics 2008; 1: 27-33.]. In the absence of HSR inducer the pathway is inactive, therefore there is little beta-lactamase expression. When the cells are loaded with CCF4-AM and are excited at 406 nm, the substrate emits at around 530 nm (green). However, when cells are stimulated with 17-AAG the pathway is activated, leading to beta-lactamase reporter gene expression. The substrate is then cleaved by beta-lactamase, thereby disrupting the fluorescence resonance energy transfer (FRET) and resulting in emission at 460 nm (blue) following excitation at 406 nm.
METHODS
Generation of the Stable HSE-bla Reporter Line
Three contiguous copies of the HSE sequence (CTGGAATATTCCCGACCTGGCAG) present in the HSP70 gene promoter was cloned into pLenti-bsd/MCS-bla vector (Invitrogen, Carlsbad, CA) via ClaI and NheI sites. Lentivirus was prepared according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA). HeLa cells were transduced with the lentivirus expressing HSE-bla and selected for blasticidin resistance. Blasticidin resistant cells were left untreated or treated with heat shock at 42°C for 30 minutes, and then returned to 37°C. Cells were allowed to recover for 4.5 hours and then loaded with CCF4-AM substrate for 2 hours at room temperature prior to collecting a pool of heat shock induced beta-lactamase expressing cells by FACS. The cells were expanded and further tested in 384-well plate format for dose response using HSR inducer 17-AAG and HSF-specific RNAi knock-down.
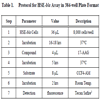
HSE-bla Assay in 384-well Plate Format
Cells in a sub-confluent state (or cryopreserved cells) were resuspended in Assay Medium (DMEM with GlutaMAXTM (Invitrogen, Carlsbad, CA, catalog number 10569) supplemented with 0.1% dialyzed FBS, 0.1 mM NEAA, 25 mM HEPES, 100 U/mL Penicillin and 100 µg/mL Streptomycin) and plated in a 384-well assay plate (Corning, Lowell, MA, catalog number 3712) at 8,000 cells (36 μL) per well (Table 1). 36 μL of Assay Medium without cells were plated in cell-free control wells on the same plate. Following overnight incubation at 37ºC, the cells were then stimulated with 4 µL/well of 10x 17-AAG (LC Laboratories, MA, catalog number A-6880) over the indicated concentration range for 5 hours before adding 8 µL/well of 6x Live-BLAzerTM-FRET B/G Substrate mixture (Invitrogen, catalog number K1096) for 2 hours. Fluorescence intensity at excitation 406 nm and emission 460 nm and 530 nm were obtained using a Tecan Safire2 fluorescence plate reader (Tecan, Durham, NC).
After subtracting the average fluorescence intensity from the cell-free control wells, the 460 nm/530 nm emission ratio was calculated. Response Ratio is a measurement of the assay window and is calculated as the 460 nm/530 nm emission ratio of the stimulated wells divided by the 460 nm/530 nm emission ratio of the unstimulated wells. Response Ratio provides a consistent way for data comparison, especially when data generated from different plates are compared and when different plate reader settings are used which usually result in large variables in the 460 nm/530 nm emission ratio. Response Ratios were plotted against test ligand concentrations in log scale and then analyzed using Prism software (GraphPad Software, Inc. San Diego, CA). A sigmoidal dose-response equation with varying slope was used to fit the data and generate EC50 values. Z’-factor values were calculated as: Z’-factor = 1 - [(3 x stdevunstim + 3 x stdevmaxstim) / (avgmaxstim – avgunstim)].
For the compound inhibition experiment, cells were seeded into a 384-well assay plate at 8,000 cells (32 μL) per well. Following overnight incubation at 37°C, cells were pretreated for 1 hour with 4 µL of quercetin (Sigma-Aldrich, Saint Louis, MO) at various final concentrations from 0.01 to 200 µM. Cells were then stimulated with 4 µL of 17-AAG at a final concentration of 65 nM (EC80) for 5 hours before the beta-lactamase assay was performed as described above.
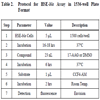
HSE-bla Assay in 1536-well Plate Format
Assay protocol is described in Table 2. Briefly, HSE-bla HeLa cells were resuspended in OPTI-MEM® I medium (Invitrogen, CA, catalog number 51895) containing 0.1% dialyzed FBS, and dispensed at 1500 cells/5 μL/well in 1536-well black wall/clear bottom plates (Kalypsys, San Diego, CA) using a Flying Reagent Dispenser (Aurora Discovery, Carlsbad, CA). After the cells were incubated at 37ºC overnight, 23 nL of DMSO or 17-AAG at concentration range of 0.1 nM to 3.83 µM was transferred to the assay plate by a pin tool (Kalypsys, San Diego, CA) resulting in a 217-fold dilution. The plates were incubated at 37°C for 6 hours. After 1 µL of LiveBLAzer™ B/G FRET substrate (Invitrogen, Carlsbad, CA) was added, the plates were incubated at room temperature for an additional 2 hours and then fluorescence intensity at 405 nm excitation and 460 nm and 530 nm emission was measured using an Envision plate reader (Perkin Elmer, Shelton, CT). Data was expressed as the ratio of 460 nm/530 nm emissions without background subtraction.
RNAi Experiment
HSE-bla HeLa cells were plated in 384-well format in Growth Medium (DMEM containing 10% dialyzed FBS, 0.1 mM NEAA and 25 mM HEPES) at 2,000 cells/well and reverse transfected using LipofectamineTM RNAiMAX Transfection Reagent (Invitrogen, Carlsbad, CA) and 20 nM of Stealth™ RNAi duplexes against HSF1, HSF2, and HSF4. RNAi oligo targeting beta-lactamase was used as the positive control and a random-sequence oligo with 48% GC content (MedGC) as the negative control. A set of 2 oligos for each HSF was used:
HSF1-1(GGAUGCUAUGGACUCCAACCUGGAU)
HSF1-2(UGCGGCAGCUCAACAUGUAUGGCUU)
HSF2-1(GCAUAGACCCAGAUCUCCUGGUUGA)
HSF2-2 (CCCUUUGGAAGGAGGUGUCAGAAUU)
HSF4-1 (GGUCAUUGGCAAGCUGAUCCAGUGU)
HSF4-2 (CCCUACUUCAUCCAGUCGCCUUCUA)
Cells were incubated with RNAi oligos at 37°C for 40 hours, followed by a medium change to Assay Medium and then stimulation with 17-AAG for 6 hours before the beta-lactamase assay was performed as described above. For heat shock treatment, 40 hours post-transfection the plate was subjected to heat shock at 42°C for 1 hour followed by a 5-hour incubation at 37°C prior to performing the beta-lactamase assay.
RESULTS AND DISCUSSION
Assay Development
To generate a cell-based assay suitable for high-throughput screening for modulators of HSR, we stably integrated an HSF-responsive beta-lactamase reporter element, HSE-bla, into the HeLa human cervical cancer cell background as detailed in Methods. Previous studies have established that HeLa cells respond to heat shock leading to the activation of HSF1 and therefore serve as a suitable cell background for interrogating this pathway [10Skaggs HS, Xing H, Wilkerson DC, et al. HSF1-TPR interaction facilitates export of stress-induced HSP70 mRNA J Biol Chem 2007; 282: 33902-7.]. Following antibiotic selection, a highly responsive pool of stable integrants was isolated that provides an effective HSR readout, with low beta-lactamase activity detected from unstimulated cells (i.e. mostly green cells) and efficient induction (i.e. mostly blue cells) observed following incubation at 42°C or by using HSP90 inhibitor 17-AAG (Fig. 1 and Fig. 2A
and Fig. 2A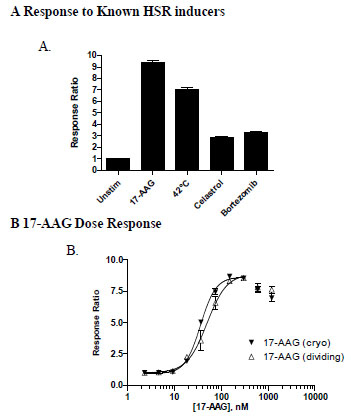 ).
).
HSE-bla HeLa cells responded to 17-AAG in a dose dependent manner with an EC50 value of 49 nM (Fig. 2B ), consistent with literature results [5Bagatell R, Paine-Murrieta GD, Taylor CW, et al. Induction of a heat shock factor 1-dependent stress response alters the cytotoxic activity of hsp90-binding agents Clin Cancer Res 2000; 6: 3312-18.]. At the highest two concentrations tested, a slight drop of response ratio was observed, presumably due to the negative feedback signal since no significant cytotoxicity was observed at these concentrations of 17-AAG. It is possible that high concentration of 17-AAG leads to high expression of HSP70 which then sequesters HSF1 from activating downstream reporter gene expression.
), consistent with literature results [5Bagatell R, Paine-Murrieta GD, Taylor CW, et al. Induction of a heat shock factor 1-dependent stress response alters the cytotoxic activity of hsp90-binding agents Clin Cancer Res 2000; 6: 3312-18.]. At the highest two concentrations tested, a slight drop of response ratio was observed, presumably due to the negative feedback signal since no significant cytotoxicity was observed at these concentrations of 17-AAG. It is possible that high concentration of 17-AAG leads to high expression of HSP70 which then sequesters HSF1 from activating downstream reporter gene expression.
The HSE-bla HeLa cells were also shown to be responsive, to a lesser extent, to additional HSR inducers such as the triterpenoid antioxidant celastrol and proteasome inhibitor bortezomib (Fig. 2A ), consistent with previously reported activities for these inducers [1Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation J Biol Chem 2005; 280: 33097-100., 3Powers MV, Workman P. Inhibitors of the heat shock response: biology and pharmacology FEBS Lett 2007; 581: 3758-69.].
), consistent with previously reported activities for these inducers [1Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation J Biol Chem 2005; 280: 33097-100., 3Powers MV, Workman P. Inhibitors of the heat shock response: biology and pharmacology FEBS Lett 2007; 581: 3758-69.].
Assay-ready cryopreserved cells have recently become an important option for HTS, which demands a large quantity of cells that can provide consistent assay results [11Zaman GJ, de Roos JA, Blomenröhr M, van Koppen CJ, Oosterom J. Cryopreserved cells facilitate cell-based drug discovery Drug Discov Today 2007; 12: 521-26.]. To further test the suitability of the HSE-bla HeLa cells for HTS applications, cryopreserved cells were thawed and immediately used for setting up the assay. A nearly super-imposable 17-AAG dose response curve (EC50 of 36 nM) to that generated with dividing cells was obtained (Fig. 2B ). The Z’ values at the maximum assay window for both dividing and cryopreserved cells were 0.83 and 0.88, respectively, indicating that cryopreserved assay-ready HSE-bla HeLa cells can be generated and applied for HTS.
). The Z’ values at the maximum assay window for both dividing and cryopreserved cells were 0.83 and 0.88, respectively, indicating that cryopreserved assay-ready HSE-bla HeLa cells can be generated and applied for HTS.
RNAi Validation
HSF1 is known to be essential for HSR induced by various stressors [1Westerheide SD, Morimoto RI. Heat shock response modulators as therapeutic tools for diseases of protein conformation J Biol Chem 2005; 280: 33097-100.]. To validate the assay and to determine which HSF is required for HSR in the HSE-bla HeLa cell line, RNAi experiments were performed where the effect of HSF-specific RNAi oligos on 17-AAG or heat shock-induced beta-lactamase reporter activity was studied (Fig. 3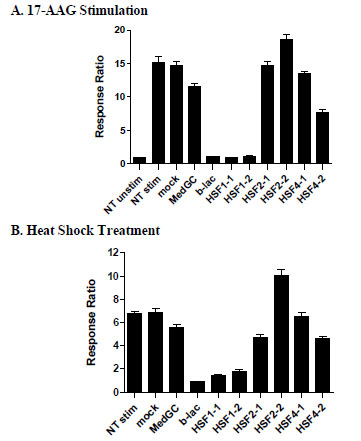 ). A beta-lactamase specific RNAi oligo (b-lac) was used as a positive control, which resulted in complete knock down of the reporter activity induced by both 17-AAG (Fig. 3A
). A beta-lactamase specific RNAi oligo (b-lac) was used as a positive control, which resulted in complete knock down of the reporter activity induced by both 17-AAG (Fig. 3A ) and 42°C heat shock (Fig. 3B
) and 42°C heat shock (Fig. 3B ). In contrast, the negative control oligo (MedGC) with random sequence and medium GC content showed minimal effects. Two RNAi oligos directed at HSF1 effectively blocked reporter activity to a similar extent as the beta-lactamase positive control under both of these HSR conditions. RNAi oligos against HSF2 and HSF4 did not show significant effect on the HSR reporter activity. These results suggest that HSF1 is essential for HSR reporter activity in HSE-bla HeLa cells, consistent with previous findings [2Voellmy R. Feedback regulation of the heat shock response Handb Exp Pharmacol 2006; 43-68.]. Moreover, these results also demonstrate the suitability of this reporter line for RNAi-mediated screening for HSR modulators.
). In contrast, the negative control oligo (MedGC) with random sequence and medium GC content showed minimal effects. Two RNAi oligos directed at HSF1 effectively blocked reporter activity to a similar extent as the beta-lactamase positive control under both of these HSR conditions. RNAi oligos against HSF2 and HSF4 did not show significant effect on the HSR reporter activity. These results suggest that HSF1 is essential for HSR reporter activity in HSE-bla HeLa cells, consistent with previous findings [2Voellmy R. Feedback regulation of the heat shock response Handb Exp Pharmacol 2006; 43-68.]. Moreover, these results also demonstrate the suitability of this reporter line for RNAi-mediated screening for HSR modulators.
Inhibitor Activity
To demonstrate screening for inhibitors of HSR signaling, dose response inhibition was performed using the flavonoid quercetin, which has been reported to inhibit HSR by down-regulating HSF1 activity [6Nagai N, Nakai A, Nagata K. Quercetin suppresses heat shock response by down regulation of HSF1 Biochem Biophys Res Commun 1995; 208: 1099-05.]. As shown in Fig. (4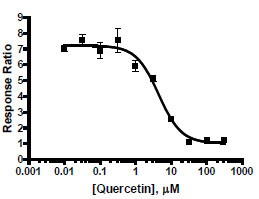 ), quercetin effectively inhibited 17-AAG treated HSE-bla HeLa cells with an IC50 of 5.8 μM, suggesting that this assay can be used to screen for HSR inhibitors.
), quercetin effectively inhibited 17-AAG treated HSE-bla HeLa cells with an IC50 of 5.8 μM, suggesting that this assay can be used to screen for HSR inhibitors.
Assay Optimization
To optimize the assay for 384-well format, several parameters were evaluated, including cell plating density, DMSO tolerance, 17-AAG stimulation time, and beta-lactamase substrate loading time. First, the effect of cell density on assay performance was tested using a range of plating densities, from 2,000 cells/well to 16,000 cells/well (Fig. 5A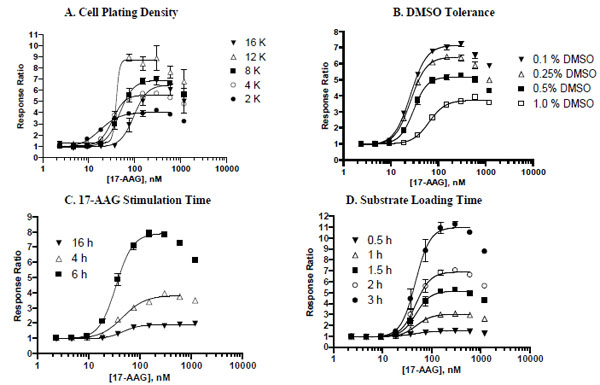 ). A robust assay window was observed with a wide range of cell plating densities, from 4,000 cells/well to 12,000 cells/well with EC50 values of 17-AAG ranging from 33 nM to 47 nM. The 8,000 cells/well plating density was selected as the basis for further assay optimization because it provided both a good assay window (7-fold) and minimal variability as exemplified by a CV (coefficient of variation) of 5% at the maximal response.
). A robust assay window was observed with a wide range of cell plating densities, from 4,000 cells/well to 12,000 cells/well with EC50 values of 17-AAG ranging from 33 nM to 47 nM. The 8,000 cells/well plating density was selected as the basis for further assay optimization because it provided both a good assay window (7-fold) and minimal variability as exemplified by a CV (coefficient of variation) of 5% at the maximal response.
To satisfy the HTS requirement for compounds to be dissolved in DMSO, a cell-based assay needs to be tolerant to certain amounts of DMSO. DMSO tolerance testing results (Fig. 5B ) indicated that the HSE-bla HeLa reporter assay can tolerate up to 0.5% final DMSO concentration with only a modest drop in assay window (from ~7.2-fold in the presence of 0.1 % DMSO to ~5.2-fold in the presence of 0.5% DMSO in this experiment). The highest concentration of DMSO (1%) tested adversely affected the reporter assay by reducing the assay window by nearly two-fold and producing a substantial right-shift in the 17-AAG EC50 value. Thus, 0.1% DMSO was consistently used in all other assay optimization experiments described here.
) indicated that the HSE-bla HeLa reporter assay can tolerate up to 0.5% final DMSO concentration with only a modest drop in assay window (from ~7.2-fold in the presence of 0.1 % DMSO to ~5.2-fold in the presence of 0.5% DMSO in this experiment). The highest concentration of DMSO (1%) tested adversely affected the reporter assay by reducing the assay window by nearly two-fold and producing a substantial right-shift in the 17-AAG EC50 value. Thus, 0.1% DMSO was consistently used in all other assay optimization experiments described here.
It usually takes a minimum of 4 to 5 hours of ligand stimulation to activate downstream gene transcription. Depending on the pathway, longer stimulation may be required for generating a robust assay window. To test the effect of 17-AAG stimulation time on assay performance, HSE-bla HeLa cells were stimulated with 17-AAG for 4, 6, or 16 hours prior to assaying beta-lactamase activity (Fig. 5C ). Among the three time points tested, a 6-hour incubation with 17-AAG resulted in the highest response with a maximal response ratio of ~7.9, nearly two-fold higher as compared to the shorter 4-hour incubation (maximal response ratio of ~4.0). Overnight incubation with 17-AAG resulted in a substantially diminished assay window (maximal response ratio of ~2.0).
). Among the three time points tested, a 6-hour incubation with 17-AAG resulted in the highest response with a maximal response ratio of ~7.9, nearly two-fold higher as compared to the shorter 4-hour incubation (maximal response ratio of ~4.0). Overnight incubation with 17-AAG resulted in a substantially diminished assay window (maximal response ratio of ~2.0).
The last parameter evaluated was the beta-lactamase substrate loading time. It is known that when the amount of beta-lactamase present in cells is low, longer substrate loading can result in more substrate conversion [8Zlokarnik G, Negulescu PA, Knapp TE, et al. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter Science 1998; 279: 84-8.]. To evaluate the effect of substrate loading time on the HSR assay outcome, an assay plate was read at various time points following addition of the substrate to the wells (Fig. 5D ). Increasing substrate loading time clearly resulted in marked increases in assay window up to a maximum of 3 hours of loading tested. Both 1.5-hour and 2-hour loading conditions generated sufficient assay windows and good Z’ values (0.80). Together a 5-hour 17-AAG stimulation and a loading time of 2 hours or shorter allows for the assay to be conveniently performed within the context of an 8-hour workday.
). Increasing substrate loading time clearly resulted in marked increases in assay window up to a maximum of 3 hours of loading tested. Both 1.5-hour and 2-hour loading conditions generated sufficient assay windows and good Z’ values (0.80). Together a 5-hour 17-AAG stimulation and a loading time of 2 hours or shorter allows for the assay to be conveniently performed within the context of an 8-hour workday.
Assay Miniaturization to 1536-well Plate Format
The HSE-bla assay was further miniaturized into 1536-well plate format with a final 5 μl assay volume. To check the assay quality the assay was tested with a DMSO plate containing a final DMSO concentration of 0.46%. In the DMSO/CV plate testing, the signal to background ratio (S/B) was 3.3 fold, CV was 6.4% and Z’ factor was 0.66 (Fig. 6A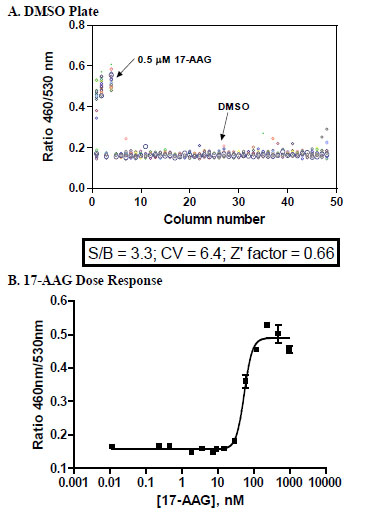 ). Fig. (6B
). Fig. (6B ) shows the dose response curve of 17-AAG that was generated in 1536-well format, yielding an EC50 of 54 nM that correlates well with the EC50 (49 nM) obtained in 384-well format. These results demonstrate that the HSE-bla assay can be run in 1536-well format and is robust and suitable for high-throughput screening.
) shows the dose response curve of 17-AAG that was generated in 1536-well format, yielding an EC50 of 54 nM that correlates well with the EC50 (49 nM) obtained in 384-well format. These results demonstrate that the HSE-bla assay can be run in 1536-well format and is robust and suitable for high-throughput screening.
CONCLUSIONS
In conclusion, we have developed a beta-lactamase reporter gene assay that can be used for high-throughput screening for modulators of the HSR pathway, which can be then deconvoluted with lower-throughput follow-up assays, such as HSP90 binding assays and/or assays monitoring the phosphorylation of HSF1. Taking together the advantages of the beta-lactamase reporter technology, which allows for a ratiometric and sensitive read-out, this reporter assay can be miniaturized to 384-well and 1536-well formats. We have demonstrated the suitability of this assay for detecting both inducers and inhibitors of HSR signaling using known small molecule modulators and HSF-specific RNAi oligos. It is anticipated that compound library screening using the HSE-bla HeLa assay will provide new leads in the search for novel modulators of HSR.
ACKNOWLEDGMENT
The authors would like to thank Pam Whitney for FACS support.