- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

Current Chemical Genomics and Translational Medicine
(Discontinued)
ISSN: 2213-9885 ― Volume 12, 2018
A Multiplexed Fluorescent Calcium and NFAT Reporter Gene Assay to Identify GPCR Agonists
Heeral Sheth 1, #, Colleen Gorey 2, #, Nicole Roush 3, Shelly Smallman 3, Elizabeth Collantes 3, Maxine Santoro 3, Barbara Olson 3, Laura Fitzgerald 3, Paul H Lee 4, Xiqiang John Shen 3, *
Abstract
Intracellular calcium response and resulting calcium signaling to an agonist-GPCR interaction are important for the measurement of compound activity in the GPCR drug development. The increase in cytosol calcium concentration can be measured by the fluorescent calcium indicator dye such as Fluo-4 in a quick assay (in 3-5 minutes) using the fluorescence imaging plate reader. The calcium signaling through the transcription factors such as NFAT that induces gene expression can be measured by the reporter gene assay that links to the expression of reporter enzyme such as the beta-lactamase that requires 5-hour incubation. We have evaluated a multiplexed assay that sequentially measures the calcium response to a GPCR agonist in a rapid fluorescent calcium dye assay, followed by a NFAT beta-lactamase assay, and compared them in the single assay format. We found that the agonist activity determined in the multiplexed assay were comparable with these determined in the single assay format and the Z’ factors were all > 0.5. Five active compounds were identified that were active in both calcium dye assay and beta-lactamase assay. Therefore, our results demonstrated the utility of this multiplexed calcium assay for screening of GPCR compounds that can cross validate the primary hits and help to eliminate the false positive compounds.
Article Information
Identifiers and Pagination:
Year: 2013Volume: 7
First Page: 1
Last Page: 8
Publisher Id: CCGTM-7-1
DOI: 10.2174/2213988501307010001
Article History:
Received Date: 21/7/2011Revision Received Date: 20/12/2012
Acceptance Date: 31/12/2012
Electronic publication date: 3/4/2013
Collection year: 2013
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Therapeutics for Rare and Neglected Diseases (TRND), National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH), 9800 Medical Center Drive, Rockville, MD 20850, USA; Tel: +1-301-827-2512; Fax: +1-301-827-2534; E-mail: john.shen@nih.gov# Both authors contribute equally
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 21-7-2011 |
Original Manuscript | A Multiplexed Fluorescent Calcium and NFAT Reporter Gene Assay to Identify GPCR Agonists | |
INTRODUCTION
G-protein-coupled receptors (GPCRs) are a large family of trans-membrane receptors that bind to extracellular agonists to activate intracellular signal transduction pathways including calcium, cAMP, ERK-phosphorylation and beta-arrestin [1Flordellis CS. The plasticity of the 7TMR signaling machinery and the search for pharmacological selectivity Curr Pharm Des 2012; 18: 145-60., 2Kimple AJ, Bosch DE, Giguere PM, Siderovski DP. Regulators of G-protein signaling and their Galpha substrates promises and challenges in their use as drug discovery targets Pharmacol Rev 2011; 63: 728-49.]. The location of these receptors on the cell membrane and the extracellular interaction site of ligands with receptors allow easily accessing of therapeutic compounds that modify the biological functions of these receptors. GPCRs are traditional targets of drug therapies and there are more than 30% of the currently available medications that are targeting GPCRs [3Hopkins AL, Groom CR. The druggable genome Nat Rev Drug Discov 2002; 1: 727-30., 4Wise A, Gearing K, Rees S. Target validation of G-protein coupled receptors Drug Discov Today 2002; 7: 235-46.].
Intracellular calcium release stimulated by a GPCR agonist initiates an important signaling pathway that triggers a variety of cellular functions including cell secretion, muscle contraction and neurotransmitter release. Calcium mobilization also activates downstream calcium-dependent transcription factors, such as nuclear factor of activated T-cells (NFAT) that regulates gene expression [1Flordellis CS. The plasticity of the 7TMR signaling machinery and the search for pharmacological selectivity Curr Pharm Des 2012; 18: 145-60.]. Because the calcium mobilization occurs early in the signaling pathway, it is a measurement point for assessing the quick response of compounds. The measurement of NFAT response on a GPCR agonist usually involves gene transcription that reports the late response of GPCR activation. Thus, a combination of an early response with a late response for a compound screening assay may help to identify additional information and property of the compounds acting on the GPCR.
Cell based assays with the high throughput screening (HTS) technology have become increasingly popular in drug discovery and development [5Hauptschein RS, Eustace BK, Jay DG. Global high-throughput screens for cellular function Exp Hematol 2002; 7: 235-46.-7Liu K, Titus S, Southall N, et al. Comparison on functional assays for Gq-coupled GPCRs by measuring inositol monophospate-1 and intracellular calcium in 1536-well plate format Curr Chem Genomics 2008; 1: 70-8.]. The new challenge in cell based assays includes generation of an appropriate cell model, adaptation to robotic screening platform, and elimination of false positive compounds. Currently, the main HTS assay for measurement of cytosol calcium concentration is based on the fluorescent calcium binding dyes such as Fluo-4 (excitation wavelength of 488 nm, emission wavelength of 520 nm). Other calcium indicator dyes are also available including Fura-2, Indo-1, Calcium Green, and Calcium Orange though they are not appropriate for HTS. The NFAT- reporter gene assay has also been used for the measurement of intracellular calcium response to a GPCR agonist such as the beta-lactamase assay with the GeneBLAzer® technology [8Oosterom J, van Doornmalen EJ, Lobregt S, Blomenrohr M, Zaman GJ. High-throughput screening using beta-lactamase reporter-gene technology for identification of low-molecular-weight antagonists of the human gonadotropin releasing hormone receptor Assay Drug Dev Technol 2005; 3: 143-54., 9Zlokarnik G, Negulescu PA, Knapp TE, et al. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter Science 1998; 279: 84-.]. In this assay, increase in intracellular calcium by a GPCR agonist stimulates the production of beta-lactamase that turns the substrate from green color to blue color (Fig. 1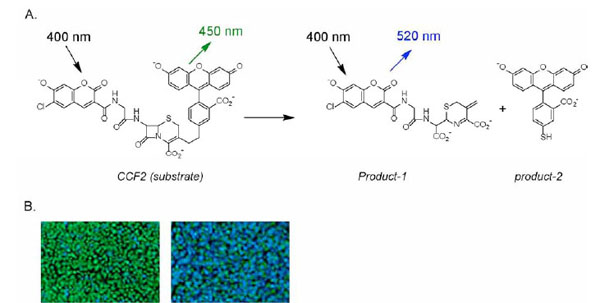 ). Here we report a multiplexing assay approach for GPCR compound screens that sequentially measures an intracellular calcium response detected by the calcium indicator Fluo-4 and the activity of beta-lactamase reporter in response to GPCR agonists or compounds. This multiplex assay enables measurements of GPCR-Ca signaling pathway at two different levels, which has not only saved time and cost for HTS but will also help to eliminate false positive hits.
). Here we report a multiplexing assay approach for GPCR compound screens that sequentially measures an intracellular calcium response detected by the calcium indicator Fluo-4 and the activity of beta-lactamase reporter in response to GPCR agonists or compounds. This multiplex assay enables measurements of GPCR-Ca signaling pathway at two different levels, which has not only saved time and cost for HTS but will also help to eliminate false positive hits.
 |
Fig. (1) Schematic diagram of coumarin cephalosporin fluorescein (CCF) cleaved by β-lactamase. After the substrate CCF-acetoxymethylester (AM) enters the cell, it is converted into CCF by endogenous esterases, thereby trapping it inside the cell.Excitation of CCF at 409 nmleads to FRET (A) from the coumarin moiety to the fluorescein derivative that produces green fluorescencedetectable at 520 nm (green cells,B). In the presence of β-lactamase, the CCF substrate is cleaved to form two fluorophores that disrupts FRET. Excitation at 409 nm now produces a fluorescence detectable at 447 nm (blue cells, B) [8Oosterom J, van Doornmalen EJ, Lobregt S, Blomenrohr M, Zaman GJ. High-throughput screening using beta-lactamase reporter-gene technology for identification of low-molecular-weight antagonists of the human gonadotropin releasing hormone receptor Assay Drug Dev Technol 2005; 3: 143-54.,9Zlokarnik G, Negulescu PA, Knapp TE, et al. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter Science 1998; 279: 84-.] |
MATERIALS AND METHODS
Materials
The CHO Gα15NFATbla-MT1 cell line stably expressing the MT1 (melatonin receptor 1) and β-lactamase under control of the NFAT response element was obtained from Donald Vanleeuwen (Pfizer Global R&D). The Fluo-4 NW (NW: no wash) Calcium Assay Kit was obtained from Molecular Probes/Life Technologies. The GeneBLAzer® assay kit was obtained from Invitrogen (Carlsbad, CA). The compounds were ordered from the Pfizer compound library. All cell culture components were obtained from Invitrogen (Carlsbad, CA), with the exception of Puromycin and Probenecid in NaOH solution, which were obtained from Sigma-Aldrich (St. Louis, MO). The assay buffer components used for the multiplexing experiment were Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen, #31053-028), dialyzed Fetal Bovine Serum (dFBS, Invitrogen #26400-036), Non-Essential Amino Acids (NEAA, Invitrogen, #11140-050), HEPES (Invitrogen, #15630-080), L-glutamine (Invitrogen, #35050-061), Na-Pyruvate (Invitrogen, #11360-070), and Probenecid in NaOH (Sigma Aldrich, P8761). Costar® (Cambridge, MA) 384-well plates with clear bottoms (#3712) were used for seeding and assaying the CHO-MT1 cells.
Cell Culture
MT1 NFAT-bla Frozen CHO cells were frozen at 3x107 cells/ml in DMEM supplemented with 10% dFBS, 0.1 mM nonessential amino acids, 25 mM HEPES, Zeocin, Blasticidin, and Puromycin. The cells were grown in T-150 flasks (BD Biosciences, #15705-074) at 37 °C/5% CO2, and frozen into vials. One vial was sufficient for seeding 5 plates at 12,000 cells per well.
Fluo-4 Calcium Dye Assay
The Fluo-4 NW dye was prepared according to the manufacturer’s directions, with the exception of the buffer medium (included in the pack) being replaced by the assay medium containing phenol-red free DMEM, 0.1% dFBS, 0.1mM NEAA, 25mM HEPES, 3.97mM L-Glutamine, 1mM NaPyruvate, and 2.5mM Probenecid in NaOH. The cells were plated overnight in a 384-well plate in phenol-red DMEM, 5% dFBS, 0.1mM NEAA, and 25mM HEPES, at 12,000 cells/well in 40μl of solution. Cells were incubated overnight at 37 °C/5% CO2 to allow the cells to adhere to the plate bottom. The following day, the media was removed from the wells, and replaced with 30μl of Fluo-4 NW dye solution. Cells were incubated at 37 °C/5% CO2 for 1 hour, and assayed immediately. The assay medium added to the Fluo-4 NW dye in the stand-alone protocol contains 90% Hank’s Balanced Salt Solution, 25mM HEPES, and 2.5mM Probenecid in NaOH solution.
Compound Plate Preparation
A set of 32 96-well master stock plates were prepared at a high concentration of 4mM, with a 10 point curve at a 1:4 dilution ratio. Two columns are high control of 2-iodomelatonin at 1mM, and two columns are low control of 100% DMSO. Each master plate was stamped into four compound assaying plates at 0.5μl per well, and diluted with 50μl of the same assay medium used to dilute the Fluo-4 NW dye. The final high concentration of testing compound and final high control were 4μM and 1μM, respectively. Final DMSO concentration was 0.25%.
Multiplexed Fluo-4 Calcium and NFAT-Beta-Lactamas Reporter Gene Assay
The FLIPR liquid handling device from Molecular Devices (Sunnyvale, CA) had a 384 well-head, and was used for compound addition and testing with Fluo-4 NW dye for the FLIPR multiplexed and stand-alone screens. During compound testing, cells were stimulated with 10uL of the compound, and control wells were stimulated with 2-iodomelatonin. Each 384-well cell plate contained a control column of wells without cells creating a background subtraction column. Data was obtained for ~180 seconds after compound addition, and plotted as the difference between the maximum and minimum stimulation values (ΔMax-Min). Tips were changed between each cell plate. For the β-lactamase stand-alone screen, compound was added to the cell plate using Molecular Devices FX robotic liquid handler. Following compound addition, the plates were incubated at 37 °C/5% CO2 for 5 hours.
Cells were then loaded with GeneBLAzer® CCF4-AM dye. For the β-lactamase multiplexed and stand-alone screens, a loading solution was prepared consisting of 12μl of solution A, 60μl of Solution B, 925μl of Solution C, and 75μl of Probenecid in Phosphate buffer. Then, 10μl of the CCF4-AM dye solution was added to each well of the screening plates. The plates were incubated at 37 °C/5% CO2 for 2 hours at room temperature (in the dark to prevent degradation by light) prior to reading fluorescence on the Analyst AD from Applied Biosystems (Foster City, CA) at an excitation filter of 425 nm and emission filters of 460/40 nm and 530/30 nm. Data are presented in blue/green ratio of each well divided by the average of the blue/green ratio of the un-stimulated control wells.
Data Analysis
Data for the Fluo-4 NW screen were analyzed in SpotFire® (TIBCO Software Inc., Palo Alto, CA), a valuable tool for removing outliers and presenting EC50 values from the linear regression model of each agonist. Microsoft Excel (Redmond, WA) was used to compile all data from the multiplexed and stand-alone screens.
The % activity was calculated for the Fluo-4 NW and β-lactamase data sets. Two sets are considered: % activity above 30%, and above 50%. The multiplexed and stand-alone results were compared to analyze assay ‘hit’ consistency.
RESULTS
Multiplexing the FLIPR Calcium Assay with NFAT Beta-Lactamase Reporter Gene Assay
The FLIPR calcium assay with a Fluo-4 no wash dye kit in the single assay format used the standard assay buffer consisting of 90% HBSS with 25mM HEPES, and 2.5mM Probenecid. However, this assay buffer was not adequate for the multiplexed assay because the cells did not survive the 5hr incubation for the subsequent NFAT beta-lactamase assay followed the FLIPR calcium assay. The growth media and overnight assay media for the MT1 CHO cells contained DMEM and dFBS. We found that the optimized assay buffer for the multiplexed assay consisted of DMEM without phenol-red, high glucose, Na-Pyruvate and L-glutamine and 0.1% dFBS with 2.5 mM Probenecid. The cells in this assay buffer produced optimal signal in the FLIPR calcium assay with the Fluo-4 no wash dye (Fig. 2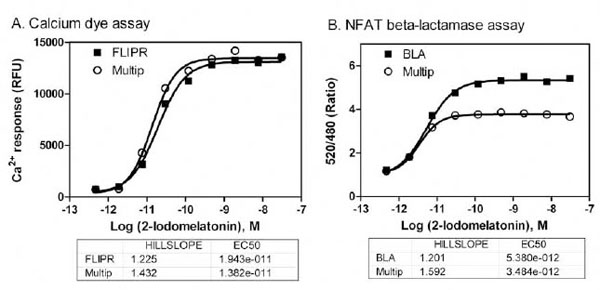 ). The slight reduction in the beta-lactamase signal in this assay buffer may be a result from the incubation with Probenecid for 5 hours. However, the EC50 values of MT1 agonist remained same in the β-lactamase assay determined in the multiplexed assay format compared with that in the single assay format.
). The slight reduction in the beta-lactamase signal in this assay buffer may be a result from the incubation with Probenecid for 5 hours. However, the EC50 values of MT1 agonist remained same in the β-lactamase assay determined in the multiplexed assay format compared with that in the single assay format.
The use of frozen cells enables preparing large amount of cells in advance and pools all the cells into the same batch, which: simplifies the HTS process, minimizes the variation due to cell passage numbers, and differences in day-to-day cell culture [10Kunapuli P, Ransom R, Murphy KL, et al. Development of an intact cell reporter gene beta-lactamase assay for G protein-coupled receptors for high-throughput screening Anal Biochem 2003; 314: 16-29.]. The direct use of resuspended frozen MT1 cells were also compared with the fresh cells for both FLIPR calcium assay and NFAT beta-lactamase assay in the single assay and multiplexed assay formatted. We found that the assay windows (signal-to-basal ratio) and EC50 values of the MT1 agonist were similar in the assays with frozen cells compared with the fresh cells. The results demonstrated the effectiveness of using frozen cells for compound screening (Fig. 3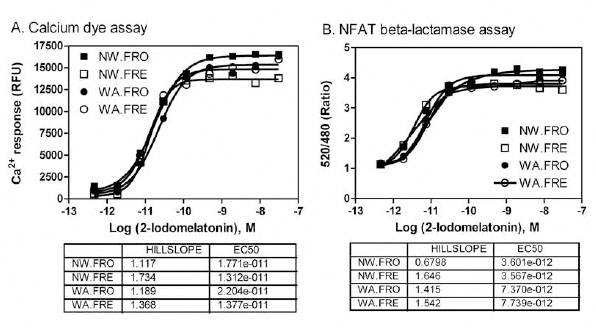 ).
).
Assay Validation
Both FLIPR calcium assays with the Fluo-4 dye and beta-lactamase assay using the MT1 CHO cells were carried out in the single assay format and multiplexed assay format to compare the EC50 values of agonist and assay performance. As illustrated in the step-by-step assay procedure for the multiplexed assay (Fig. 4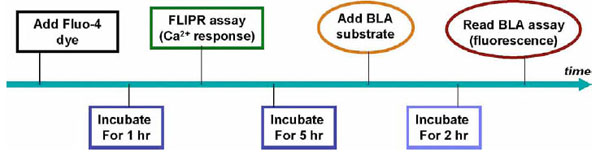 ), cells in the assay plates, after one day culture, were incubated with Fluo-4 dye for one hour followed by the measurement of intracellular calcium response to the MT1 agonist in the standard FLIPR assay. After an additional 5 hr incubation with the MT1 agonist for the induction of the beta-lactamase reporter gene, the beta-lactamase substrate was added to the assay plates and followed by an additional 2 hr incubation. The beta-lactamase activity was then measured by detecting emissions at 465 and 520 nm from excitation at 409 nm. Thus, both assays were measured sequentially from the same assay plate.
), cells in the assay plates, after one day culture, were incubated with Fluo-4 dye for one hour followed by the measurement of intracellular calcium response to the MT1 agonist in the standard FLIPR assay. After an additional 5 hr incubation with the MT1 agonist for the induction of the beta-lactamase reporter gene, the beta-lactamase substrate was added to the assay plates and followed by an additional 2 hr incubation. The beta-lactamase activity was then measured by detecting emissions at 465 and 520 nm from excitation at 409 nm. Thus, both assays were measured sequentially from the same assay plate.
 |
Fig. (4) A step by step illustration of assay procedure for the multiplexed FLIPR calcium and NFAT beta-lactamase assay |
Primary Screening of a Compound Library
The compound screens were performed in the multiplexed assay format in comparison with these in the single assay format. We found that all Z’ factor values were greater than 0.5 in the screening test determined in the both assay in the single or multiplexed assay formats (Fig. 5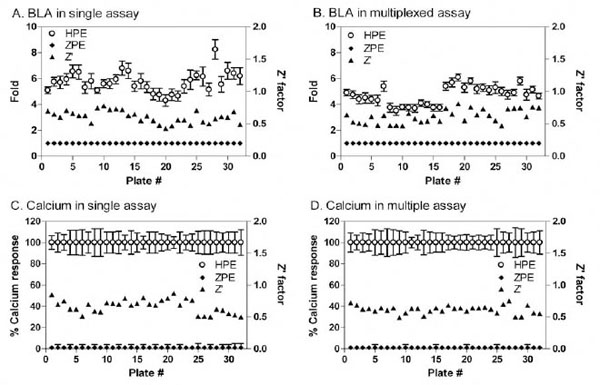 ). The assay performance was robust in all the assay formats. The EC50 values and % activities for each screening format were comparable. The Fluo-4 multiplex and stand-alone formats had corresponding maximum fluorescence outputs; however, the blue/green ratio of the β-lactamase multiplexed format was slightly lower than the stand-alone protocol. This may be due to the low dFBS concentration in the assay buffer, or the addition of Probenecid (needed for the FLIPR calcium assay)
). The assay performance was robust in all the assay formats. The EC50 values and % activities for each screening format were comparable. The Fluo-4 multiplex and stand-alone formats had corresponding maximum fluorescence outputs; however, the blue/green ratio of the β-lactamase multiplexed format was slightly lower than the stand-alone protocol. This may be due to the low dFBS concentration in the assay buffer, or the addition of Probenecid (needed for the FLIPR calcium assay)
for 5 hr incubation in the beta-lactamase assay. However, the EC50 values of the compounds were similar in the multiplexed assay compared with these in the single assay format of NFAT beta-lactamase assay (Fig. 6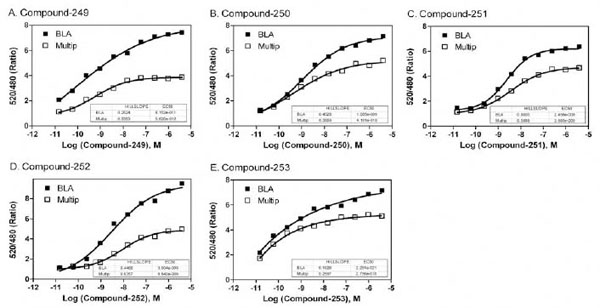 ), indicating the results were comparable in both assays.
), indicating the results were comparable in both assays.
A total of 253 compounds were selected as the primary hits and the hit rate of ~3 % at a cut-off of >30% activity or ~1.7 % at >50% activity. Of these hits, 16 compounds were active in all four assay formats (>30% activity) including two in the multiplexed assay and two in the single assay format. For the beta-lactamase assay, there was a 70% correlation of these hits between the multiplexed assay format and single assay format, while an 85% correlation of the hits was observed for the FLIPR calcium assay in both assay formats. On the other hand, the multiplexed assay format showed a 73% hit rate correlation between the beta-lactamase assay and the FLIPR calcium assay (Tables 1, 2, and 3).
The EC50 values for each of the 16 hits that were active across all four assay formats are shown in Table 4. Five compounds of interest to the MT1 project were also potent hits in all four assay formats (Fig. 6 ) that were promising as potential agonists of the MT1 receptor.
) that were promising as potential agonists of the MT1 receptor.
DISCUSSION
The fluorescent calcium dye assay measures the quick calcium response to the binding of an agonist to a GPCR, while the NFAT beta-lactamase reporter gene assay measures the calcium signaling from GPCR activation then transactivation of the NFAT reporter gene, beta-lactamase. Thus, the calcium dye assay only shows the early events of GPCR activation including agonist-GPCR binding, activation of Gq protein and PLC that release second messengers IP3 and DAG. The calcium is then released by the activation of IP3 channels in the intracellular storage site ER. On the other hand, a more complete intracellular calcium signaling pathway is needed for the activation of beta-lactamase transcription by the NFAT reporter gene. Once the cytosol calcium is increased by an agonist-GPCR interaction, calcium ions bind to calmodulin, a calcium sensor protein, which activates calcineurin, a serine/threonine phosphatase and rapidly dephosphorylates NFAT proteins. This allows NFAT enters nucleus where it binds to specific promoters and initiates gene transcription. The disadvantage of NFAT beta-lactamase assay is that it may produce a large amount of false positive hits in compound screens since so many other proteins and factors are involved in its signaling. However, a desired true GPCR activator should activate both of these mechanisms. Therefore, multiplexing the calcium dye assay with the NFAT beta-lactamase reporter gene assay enables a measurement of compound activities in two assays from the same cells. It also greatly saves the screening cost and time.
Since the calcium dye assay only takes a short time, including a 1 hour dye incubation and 3 to 5 minutes for agonist response recording, a simple physiological salt buffer is usually used for the experiment. The NFAT beta-lactamase
assay requires long agonist incubation time (usually 5 hours) for the expression of reporter gene and thus cell culture medium over buffer is preferred. Thus, we have optimized the assay buffer for this multiplexed assay that consists of 0.1% dFBS, high glucose DMEM without phenol-red and with added Na-Pyruvate and L-glutamine, and 2.5 mM Probenecid. The addition of Probenecid is needed to prevent dye leaking out of cells through membrane transporters. The main advantages of using a single plate for two calcium assays for compound screens include savings in screening cost and time and more comparable results from two assays because the compound activities were determined from the same cells for the calcium dye and NFAT beta-lactamase reporter gene assays.
Cell culture to produce a large amount of cells for compound screens in a HTS format is a challenge because the day-to-day variation of yields and different cell passage numbers can cause increased variation in the screening results. Application of frozen cells has been used to minimize such a variation among the compound screening runs in different days. The frozen calls can be pooled for use in HTS that effectively minimizes the passage variations. The readily availability of frozen cells that have been stored in a freezer in aliquots allows compound screens in HTS formats without being limited by the cell culture that is usually a bottleneck in cell based screens.
In conclusion, we have developed and validated a multiplexed GPCR-calcium response assay that sequentially measures agonist activities in a rapid calcium dye assay followed by a NFAT beta-lactamase reporter gene assay. The compound activities are determined in the same cells in a single assay plate for both calcium assays, resulting in more comparable results for the same compound. It also significantly saves the screening time and cost of reagents. Therefore, this multiplex calcium assay is a useful tool for GPCR screens of large size compound libraries in the drug discovery process.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
ACKNOWLEDGEMENTS
The authors acknowledge C.K. Chan, Sally Daniels, Christopher Haber, Eric Nordby, Benjamin Turner, Jurgen Vanhauwe, Rathna Iyer, Donald Vanleeuwen, Kevin Wass, Scott Wise, and Jeff Scholten.