- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Different Isoforms of HPV-16 E7 Protein are Present in Cytoplasm and Nucleus
H Valdovinos-Torres1, M Orozco-Morales1, A Pedroza-Saavedra1, L Padilla-Noriega3, F Esquivel-Guadarrama2, L Gutierrez-Xicotencatl*, 1
Abstract
The E7 protein of high risk HPV types has been found with different molecular weights, mainly because of phosphorylation, an event that changes protein charge and mobility in SDS-PAGE. Distribution of E7 protein in the cellular compartments has also been subject of debate as some groups report the protein in nucleus and others in cytoplasm. The different subcellular distribution and molecular weights reported for the E7 protein suggest the presence of isoforms. We examined this possibility by using several antibodies that recognize different epitopes on the HPV-16 E7 protein. We showed that E7 is processed in 3 isoforms with different molecular weights and isoelectric points (IEP), and described as E7a1 (17.5 kDa, IEP 4.68), E7a (17 kDa, IEP 6.18) and E7b (16 kDa, IEP 6.96). The immunofluorescense results also showed that E7 is distributed into different compartments (ER, Golgi and nucleus), which suggest the presence of other posttranslational modifications, besides phosphorylation.
Article Information
Identifiers and Pagination:
Year: 2008Volume: 2
First Page: 15
Last Page: 23
Publisher Id: TOVJ-2-15
DOI: 10.2174/1874357900802010015
Article History:
Received Date: 29/1/2008Revision Received Date: 14/2/2008
Acceptance Date: 5/3/2008
Electronic publication date: 26/3/2008
Collection year: 2008
open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.5/), which permits unrestrictive use, distribution, and reproduction in any medium, provided the original work is properly cited.
* Address correspondence to this author at the Research Center of Infectious Diseases, National Institute of Public Health, Av. Universidad 655, Santa Ma. Ahucatitlan, Cuernavaca, Mor, Mexico CP 62508; Tel: (52) 777-3-29-30-86; Fax: (52) 777-317-54-85; E-mail: mlxico@correo.insp.mx
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 29-1-2008 |
Original Manuscript | Different Isoforms of HPV-16 E7 Protein are Present in Cytoplasm and Nucleus | |
INTRODUCTION
Cervical cancer (CC) is one of the most prevalent diseases worldwide. CC is the second major cause of death in women from underdeveloped countries [1Parkin DM, Bray F. The burden of HPV-related cancers Vaccine 2006; 24((Suppl 3)): S11-25.] and the main etiological factor is the human papillomavirus (HPV) persistent infection status. Over 90% of malignant carcinomas of the genital tract have HPV DNA sequences; the majority of which contain the high risk HPV-16 and 18 types which are linked to the progression of cervical lesions and CC [2zur Hausen H. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis J Natl Cancer Inst 2000; 92(9): 690-8., 3Syrjänen K. Mechanisms and predictors of high-risk human papillomavirus (HPV) clearance in the uterine cervix Eur J Gynaecol Oncol 2007; 28(5): 337-51.].
The mechanisms through which HPVs induce cell transformation have been intensively investigated recently. The most abundant viral transcripts in tumor and tumor cell lines come from the E6 and E7 open reading frames that are known to be oncogenic. These two genes from HPV are necessary and sufficient to induce HPV-mediated transformation of murine cells [4Yasumoto S, Burkhardt AL, Doniger J, DiPaolo JA. Human papillomavirus type 16 DNA-induced malignant transformation of NIH 3T3 cells J Virol 1986; 57: 572-7.], immortalize human fibroblasts [5Pirisi L, Yasumoto S, Feller M, Doniger J, DiPaolo JA. Transformation of human fibroblasts and keratinocytes with human papillomavirus type 16 DNA J Virol 1987; 61: 1061-6.] and in cooperation with ras to transform baby rat kidney cells and primary human keratinocytes [6Matlashewski G, Schneider J, Banks L, Jones N, Murray A, Crawford L. Human papillomavirus type 16 DNA cooperates with activated ras in transforming primary cells EMBO J 1987; 6: 1741-6., 7Phelps WC, Yee CL, Münger K, Howley PM. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A Cell 1988; 53: 539-47.].
The transformation capacity of E7 has been shown to be limited to the high risk HPV types. The E7 protein is a 98 amino acid phosphoprotein with a predicted molecular weight of 11 kDa, however this protein shows abnormal electrophoretic mobility in SDS-PAGE gels that generates different molecular weights from 14 to up to 21 kDa, as reported by different groups [8Gage JR, Meyers C, Wettstein FO. The E7 proteins of the nononcogenic human papillomavirus type 6b (HPV-6b) and of the oncogenic HPV-16 differ in retinoblastoma protein binding and other properties J Virol 1990; 64: 723-30.-10Greenfield I, Nickerson J, Penman S, Stanley M. Human papillomavirus type 16 E7 protein is associated with the nuclear matrix Proc Natl Acad Sci USA 1991; 88: 11217-21.]. It has been suggested that the different molecular weights of E7 and its mobility in SDS-PAGE gels are the result of the acidic sequence localized in the aminoterminal region of the protein, together with its high hydrophobicity and high capacity to form oligomers [11Armstrong DJ, Roman A. The anomalous electrophoretic behavior of the human papillomavirus type 16 E7 protein is due to the high content of acidic amino acid residues Biochem Biophys Res Commun 1993; 192: 1380-7.-13Alonso LG, García-Alai MM, Nadra AD, et al. High-Risk (HPV16) Human Papillomavirus E7 oncoprotein is highly stable and extended, with conformational transitions that could explain its multiple cellular binding partners Biochemistry 2002; 41: 10510-8.].
Studies in high risk HPV types showed that E7 deregulates the cell cycle mainly by binding to and promoting degradation of the tumor suppressor retinoblastoma protein (pRb) [14Caldeira S, Dong W, Tommasino M. Analysis of E7/Rb associations Methods Mol Med 2005; 119: 363-79., 15Collins AS, Nakahara T, Do A, Lambert PF. Interactions with pocket proteins contribute to the role of human papillomavirus type 16 E7 in the papillomavirus life cycle J Virol 2005; 79: 14769-80.], resulting in the dissociation of pRb from E2F transcription factors and the premature cell progression into the S-phase of the cell cycle. In a similar way, E7 affects the pRb-related pocket proteins p107 and p130 [16Davies R, Hicks R, Crook T, Morris J, Vousden K. Human papillomavirus type 16 E7 associates with a histone H1 kinasa and with p107 through sequences necessary for transformation J Virol 1993; 67: 2521-8., 17Zhang B, Chen W, Roman A. The E7 proteins of low- and high-risk human papillomaviruses share the ability to target the pRB family member p130 for degradation Proc Natl Acad Sci USA 2006; 103: 437-42.]. It has also been demonstrated that a phosphorylated E7 isoform is able to associate with the histone H1 kinase in the late G2/M phase of the cell cycle [16Davies R, Hicks R, Crook T, Morris J, Vousden K. Human papillomavirus type 16 E7 associates with a histone H1 kinasa and with p107 through sequences necessary for transformation J Virol 1993; 67: 2521-8.]. Other studies have demonstrated that E7 is capable of inducing the enzymatic activity of the α-glucosidase in the cytoplasm which degrades intracellular glucose and provides energy for cell growth and division [18Zwerschke W, Mannhardt B, Massimi P, et al. Allosteric activation of acid α-glucosidase by the human papillomavirus E7 protein J Biol Chem 2000; 275: 9534-41.].
The different activities of the E7 protein, the heterogeneous distribution in different cellular compartments (nucleus and cytoplasm) as well as the different molecular weights reported suggest the presence of different isoforms of E7. We examined this possibility by using polyclonal and monoclonal antibodies that recognize different epitopes on the HPV-16 E7 protein. We showed that E7 is processed into 3 forms (E7a1, E7a and E7b), one of which is phosphorylated. The molecular weight and the isoelectric point (IEP) as well as the cellular localization of the E7 isoforms were different. These results demonstrate that there are different isoforms of E7 in different cellular compartments and this may be important for interaction of the E7 protein with target proteins to generate the cellular transformation.
MATERIALS AND METHODOLOGY
Antibodies.
The monoclonal antibodies (mAbs) D11, B4, C2, A5 and G7 were prepared and characterized in our laboratory [19Orozco-Morales M, Esquivel-Guadarrama F, Valdovinos-Torres H, Gutierrez-Xicotencatl L. Characterization of mouse hybridomas specific for E7 of HPV-16 In preparation]. Briefly, 20 µg of purified recombinant cII-E7 protein from HPV-16 (expressed and purified from E. coli) were used to immunize mice 3 times every two weeks. Hybridomas were screened and selected by ELISA against a different recombinant E7 protein (MS2-E7) [20Pedroza-Saavedra A, Cruz A, Esquivel F, et al. High prevalence of serum antibodies to Ras and type 16 E4 proteins of human papillomavirus in patients with precancerous lesions of the uterine cervix Arch Virol 2000; 145: 603-23.]. Further characterization was performed by immunoblot, immunoprecipitation and immunofluorescence.
The anti-E7 (C24 and C89) polyclonal antibodies were prepared in our laboratory against the cII-E7 protein purified by electro-elution. Briefly, rabbits were immunized 3 times with 50 µg of recombinant E7 protein and tested for the presence of specific E7 antibodies in the same way as mAbs.
The anti-E7 (clone ED17, C20), the anti-calnexine (clone H-70) and the anti-p21 (C-29) were from Santa Cruz Biotechnology (Santa Cruz, CA. USA). The anti-GM130 (clone 35) was from BD Biosciences (San Jose, CA. USA).
In Vitro Cell Culture Conditions.
CaSki cell line (human cervical epidermoid carcinoma, naturally HPV-16 transformed), HaCaT cells (normal human keratinocytes) and Cos-7 (kidney monkey, SV40 transformed fibroblasts) were cultured in Dulbecco's Modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and maintained in a humidified atmosphere of 5% CO2/95% air, at 37°C.
In Vivo Cell Labeling.
Cells were labeled with 50 μCi/ml [35S]-methionine and [35S]-cysteine (Promix, > 1000 Ci/mmol, GE Healthcare, Piscataway, NJ. USA), in methionine and cysteine free medium and 5% of dialyzed FBS in overnight labeling experiments or with 250 μCi/ml [35S]-methionine-cysteine for 15 min in pulse-chase experiments.
Cells labeled with [32P]-orthophosphate were starved in 0.2% FBS for 48 h and then placed in phosphate free medium for further 2 h, as described previously [21Massimi P, Pim D, Kühne C, Banks L. Regulation of the human papillomavirus oncoproteins by differential phosphorylation Mol Cell Biochem 2001; 227: 137-44.]. After this time, cells were incubated with 100 μCi/ml [32P]-orthophosphate (3000 Ci/mmol, GE Healthcare, Piscataway, NJ. USA) for 1 h (time zero). The remaining cells were incubated with 10% FBS for different times and labeled for 1 h with orthophosphate, as above, before cells were collected and lysed for immunoprecipitation as described below.
Transient Transfection of HPV-16 E7 in Cos-7 Cells.
Cos-7 cells were transfected using the DEAE-dextran method as described previously [22Hancock JF, Marshall CJ, McKay IA, et al. Mutant but not normal p21 ras elevates inositol phospholipid breakdown in two different cell systems Oncogene 1988; 3: 187-93.]. Cells were seeded on 8 well Multitest Slide (MP Biomedical, Solon, OH. USA) and grown up to 80% confluence. Cells were transfected with 5 μg of purified pcDNA or pcDNA-E7 plasmid that contains the nucleotides 562 to 858 from HPV-16 which encodes for the E7 protein (GenBank access AF 477385) (a gift from Dr. J. Berumen, Genomic Medicine Laboratory, Hospital General, Mexico D.F.). Cells were grown for different periods of time (0, 2, 4, 8, 16, 24, 48 and 72 h) after which the cells were fixed with p-formaldehyde and treated for immunofluorescence, as described below.
Immunoprecipitation and SDS-PAGE.
Cells were lysed and immunoprecipitated as described elsewhere [23Magee AI, Gutierrez L, McKay IA, Marshall CJ, Hall A. Dynamic fatty acylation of p21N-ras EMBO J 1987; 6: 3353-7.]. Briefly, cell monolayers were lysed in radioimmunoprecipitation assay (RIPA) buffer in the presence of protease inhibitors (Complete mini, Roche Diagnostics, Indianapolis, IN. USA). Cell lysates were cleared by centrifugation, and supernatants immunoprecipitated for 16 h at 4oC with 3 μg of purified IgG anti-E7 mAbs or 30 μl of anti-E7 rabbit polyclonal antibodies. The antigen-antibody complex was immunoprecipitated with Protein A-Sepharose (GE Healthcare,Piscataway, NJ. USA) for 2 h at 4oC. Rabbit anti-mouse coated Protein A-Sepharose was used for mAbs. The immunoprecipitates were dissolved in Laemmli loading buffer containing 100 mM dithiothreitol, followed by electrophoresis in a 15% SDS-polyacrylamide gel. Gels were treated for fluorography with the Enlighting reagent (NEN Life Sciences Products, Boston, MA. USA) and specific bands detected by exposing the gels to X-OMAT film.
Immunoblot.
Proteins from different cell lines were extracted in RIPA buffer, as described previously [24Brugge JS, Erikson RL. Identification of a transformation-specific antigen induced by an avian sarcoma virus Nature 1977; 269: 346-8.], in the presence of protease inhibitors (Roche Diagnostics, Indianapolis, IN. USA). Total protein extracts (1 mg/ml) were incubated with the C24 polyclonal antibody and processed for immunoprecipitation as mentioned before. Immunoprecipitates were separated on 15% SDS-PAGE gels and transferred to nitrocellulose Protean membranes (0.45µm, Whatman International Ltd., Middlesex, UK), as described by Towbin and coworkers [25Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications Proc Natl Acad Sci USA 1979; 76: 4350-4.]. The membranes were blocked with PBS-0.5% Tween containing 10% skim milk for 30 min at room temperature. Subsequently, the membranes were incubated with a 1:200 dilution of anti-E7 polyclonal antibodies or with a 1:100 dilution of anti-E7 mAbs in PBS-0.5% Tween and 5% skim milk, overnight. Blots were washed with PBS-0.5% Tween followed by incubation for 1 h under the same conditions with the secondary goat anti-rabbit or rabbit anti-mouse IgG antibodies conjugated with horseradish peroxidase (DAKO, Carpinteria, CA. USA). The membranes were developed according with the chemiluminescence kit of Perkin Elmer (Waltham, Massachusetts, USA) manufacturer instructions. Membranes were exposed to X-OMAT film.
Indirect Immunofluorescence.
Cells at 80% confluence were rinsed with PBS and fixed with 4% p-formaldehyde in PBS at room temperature for 20 min followed by treatment with permeabilization buffer [1% BSA in PBS containing either 3% Triton X-100 (for nuclear structures) or 0.2% saponine (for cytosolic and internal membranes)] for 20 min at 4oC. Subsequently, cells were incubated with anti-E7 antibodies (serum diluted 1:100 for anti-E7 polyclonal Abs and 2 ng/μl of purified IgG for anti-E7 mAbs in permeabilization buffer) for 16 h at 4oC. Slides were rinsed with PBS and incubated for 2 h at 4oC with anti-rabbit IgG conjugated with Alexa 488 (green) or anti-mouse IgG conjugated with Alexa 594 (red) (dilution 1:250 and 1:800, respectively, Molecular Probes, Carlsbad, CA. USA). Specimens were mounted in 50% glycerol in PBS and visualized under the Confocal Microscope 510 META (Carl Zeiss, Massachusetts, USA) under the Plan-Neofluor 100X /1.3 oil Ph3 lent.
2-dimensional Polyacrylamide Gel Electrophoresis (2D Gels).
Immunoprecipitated E7 protein was treated for isoelectrofocusing (IEF), as described previously [26Gutierrez L, Magee AI, Marshall CJ, Hancock JF. Post-translational processing of p21ras is two-step and involves carboxyl-methylation and carboxy-terminal proteolysis EMBO J 1989; 8: 1093-8.] with some modifications. Briefly, E7 immunoprecipitates were dissolved in IEF sample buffer (9.5 M urea, 2% NP40, 2% ampholines 3-10, 100 mM DTT). IEF used 7% ampholines (GE Healthcare, NJ. USA) pH 3-9 in 2.5 mm diameter polyacrylimide gel tubes and fixed to the adaptor for slap gels electrophoresis apparatus (BRL Wrightsville, PA. USA). The IEF was carried for 16 h at 400 V and electrophoresis for the second dimension was run as described before. The gels were treated for fluorography as described previously.
RESULTS
Characterization of HPV-16 E7 Protein with Different Antibodies.
It has been reported that E7 protein shows different molecular weights and that the phosphorylation process is probably one of the causes for these differences. Thus, we decided to identify the different forms reported using polyclonal and monoclonal antibodies, produced in our laboratory that can differentiate the various isoforms. To this end, CaSki cells were labeled with [35S]-methionine-cysteine for 16 h and the E7 protein immunoprecipitated with the different antibodies as described in Materials and Methodology. The result in Fig. (1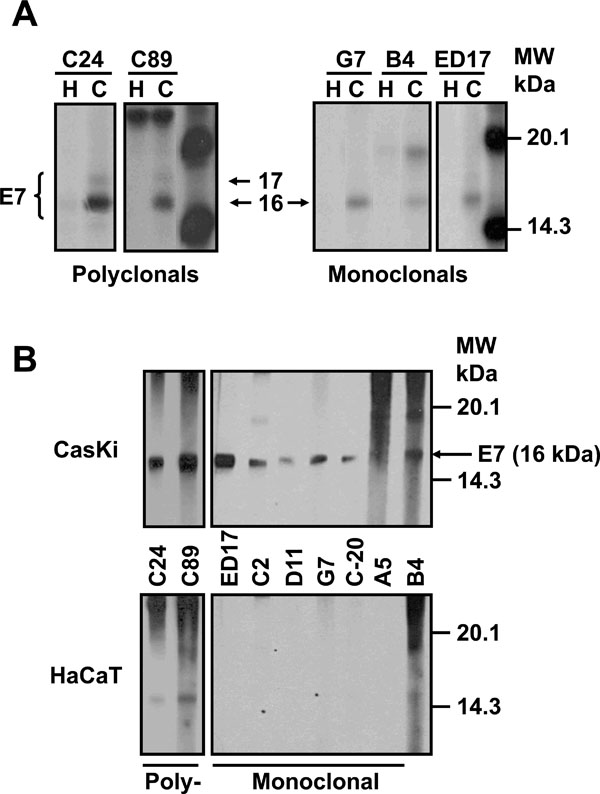 ) shows an E7 protein of 16 kDa that was immunoprecipitated with the polyclonal antibodies C24 and C89, as well as with the mAbs G7, B4 and the commercial ED17. However, a 17 kDa E7 protein was observed only with the polyclonal antibodies (C24 and C89) (Fig. 1A
) shows an E7 protein of 16 kDa that was immunoprecipitated with the polyclonal antibodies C24 and C89, as well as with the mAbs G7, B4 and the commercial ED17. However, a 17 kDa E7 protein was observed only with the polyclonal antibodies (C24 and C89) (Fig. 1A ). In contrast, none of these two bands were observed in immunoprecipitates from HaCaT cells that were used as negative controls (Fig. 1A
). In contrast, none of these two bands were observed in immunoprecipitates from HaCaT cells that were used as negative controls (Fig. 1A ), or with control rabbit or mouse serum (data not shown).
), or with control rabbit or mouse serum (data not shown).
We also tested the capacity of the antibodies to recognize the E7 protein by Western blot as the different molecular weights of the E7 protein could be the result of the different techniques used. Previously, we have shown that the sensitivity of the direct Western blot for E7 is low [27Escalante AD, Recillas TF, Valencia C, et al. Expression of E6 and E7 papillomavirus oncogenes in the outer root sheath of hair follicles extends the growth phase and bypasses resting at telogen Cell Growth Diff 2000; 11: 527-39.] and therefore, it is necessary to carry out an immunoprecipitation–Western blot to visualize the E7 protein from cell extracts. In this way, cells were immunoprecipitated first with the polyclonal C24 antibody as this antibody recognized 2 forms of E7 protein. The precipitates were separated on a SDS-PAGE gel, transferred to Protean membranes and tested with the different mAbs as described previously. The results in Fig. (1B ) showed that 5 of the 6 mAbs tested recognized the 16 kDa band of the E7 protein. This band was also recognized by the commercial mAb ED17 acquired from Santa Cruz, which was used as a positive control. The recognition of 16 kDa band is specific, as this was not present when the mAbs were tested with the HaCaT immunoprecipitates used as negative controls (Fig. 1B
) showed that 5 of the 6 mAbs tested recognized the 16 kDa band of the E7 protein. This band was also recognized by the commercial mAb ED17 acquired from Santa Cruz, which was used as a positive control. The recognition of 16 kDa band is specific, as this was not present when the mAbs were tested with the HaCaT immunoprecipitates used as negative controls (Fig. 1B ). In the case of the polyclonal antibodies (C24 and C89), they only recognized the 16 kDa band but not the 17 kDa band in the Western blot. It is possible that these antibodies only recognize a conformational epitope in the E7 protein that is lost by denaturation during the Western blot process (Fig. 1B
). In the case of the polyclonal antibodies (C24 and C89), they only recognized the 16 kDa band but not the 17 kDa band in the Western blot. It is possible that these antibodies only recognize a conformational epitope in the E7 protein that is lost by denaturation during the Western blot process (Fig. 1B ).
).
Processing of HPV-16 E7 Protein.
Processing of E7 protein was determined by pulse-chase labeling of CaSki cells and immunoprecipitated with C89 polyclonal antibody (Fig. 2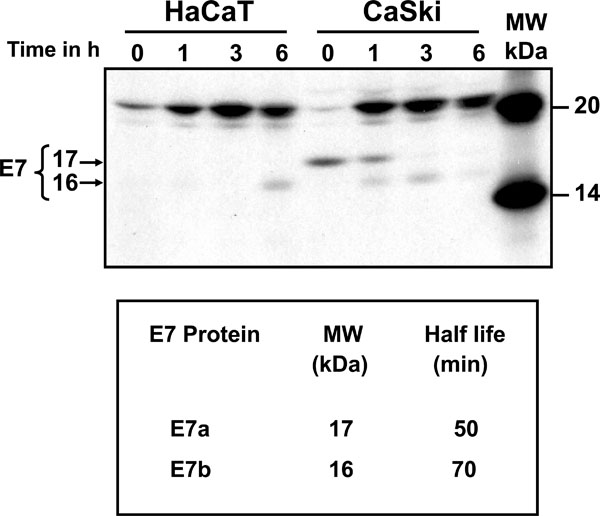 ). The top panel of Fig. (2
). The top panel of Fig. (2 ) shows the electrophoretic analysis of immunoprecipitates of [35S] methionine-cysteine labeled E7 protein after 15 min pulse, followed by chase of up to 6 h. It is clear that the E7 protein was synthesized after the 15 min pulse as a 17 kDa protein (E7a) that remained stable for up to 1 h and subsequently disappeared (Fig. 2
) shows the electrophoretic analysis of immunoprecipitates of [35S] methionine-cysteine labeled E7 protein after 15 min pulse, followed by chase of up to 6 h. It is clear that the E7 protein was synthesized after the 15 min pulse as a 17 kDa protein (E7a) that remained stable for up to 1 h and subsequently disappeared (Fig. 2 ). After 1 h of chase the E7a protein was processed to a 16 kDa band (E7b) and was stable for up to 3 h of chase (Fig. 2
). After 1 h of chase the E7a protein was processed to a 16 kDa band (E7b) and was stable for up to 3 h of chase (Fig. 2 ). This pattern of bands was not observed in the immunoprecipitates of labeled HaCaT cells used as control (Fig. 2
). This pattern of bands was not observed in the immunoprecipitates of labeled HaCaT cells used as control (Fig. 2 ). The 15 kDa band observed in HaCaT cells at 6 h (smaller than the one observed for E7 in CaSki cells) is a non-specific band as it is not consistently observed in experiment repetitions. The bands of the fluorography were scanned to calculate the half-life of the E7 proteins according to Belle and coworkers [28Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK. Quantification of protein half-lives in the budding yeast proteome Proc Natl Acad Sci U S A 2006; 103: 13004-9.]. The results showed that the half-life of the E7a protein was only 50 min and 70 min for the faster moving band of E7b (Fig. 2
). The 15 kDa band observed in HaCaT cells at 6 h (smaller than the one observed for E7 in CaSki cells) is a non-specific band as it is not consistently observed in experiment repetitions. The bands of the fluorography were scanned to calculate the half-life of the E7 proteins according to Belle and coworkers [28Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK. Quantification of protein half-lives in the budding yeast proteome Proc Natl Acad Sci U S A 2006; 103: 13004-9.]. The results showed that the half-life of the E7a protein was only 50 min and 70 min for the faster moving band of E7b (Fig. 2 , lower panel). The presence of a nonspecific 21 kDa band in pulse-chase cells (HaCaT and CaSki) was due to the high levels of label used in this kind of experiments.
, lower panel). The presence of a nonspecific 21 kDa band in pulse-chase cells (HaCaT and CaSki) was due to the high levels of label used in this kind of experiments.
Recognition of 3 Different forms of HPV-16 E7 Protein by Differences in Charge.
The pulse-chase experiments showed that the E7a protein was processed to a faster moving form described as E7b. At this point, it was not clear if there was an actual chemical modification of the protein, or if the differences were only due to a conformational change. For this reason, it was important to determine if there was any difference in charge in addition to the difference in molecular weight between these 2 isoforms of E7. Isoelectrofocusing of immunoprecipitates (C89 antibody) of overnight [35S] methionine-cysteine labeled E7 proteins was performed followed by SDS-PAGE gels. The separation of the E7 proteins in a pH gradient showed that E7a and E7b have different net charge (Fig. 3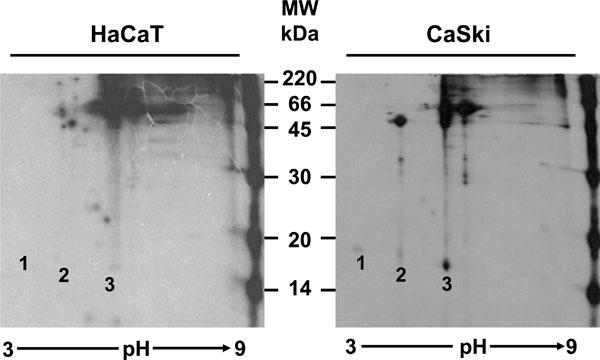 , CaSki, spots 2 and 3 respectively). However, unexpectedly an additional form of the E7 protein in the extract of CaSki cells that was not resolved in one dimension gels was found. The third band observed (E7a1) showed a molecular weight of 17.5 kDa (Fig. 3
, CaSki, spots 2 and 3 respectively). However, unexpectedly an additional form of the E7 protein in the extract of CaSki cells that was not resolved in one dimension gels was found. The third band observed (E7a1) showed a molecular weight of 17.5 kDa (Fig. 3 , CaSki, spot 1). The IEP for each form of E7 protein was calculated as 4.68 for E7a1 (spot 1), 6.18 for the E7a form (spot 2) and 6.96 for E7b (spot 3). This difference in charge among the 3 isoforms of E7 protein, suggests that the proteins are modified in some ways in two different steps.
, CaSki, spot 1). The IEP for each form of E7 protein was calculated as 4.68 for E7a1 (spot 1), 6.18 for the E7a form (spot 2) and 6.96 for E7b (spot 3). This difference in charge among the 3 isoforms of E7 protein, suggests that the proteins are modified in some ways in two different steps.
Phosphorylation of the HPV-16 E7 Protein.
Previously, it has been reported that posttranslation phosphorylation occurs in E7 [29Barbosa MS, Edmonds C, Fisher C, Schiller JT, Lowy DR, Vousden KH. The region of the HPV E7 oncoprotein homologous to adenovirus E1a and SV40 large T antigen contains separate domains for Rb binding and casein kinase II phosphorylation EMBO J 1990; 9: 153-60.]. Accordingly, we decided to identify which of the E7 forms was phosphorylated. To this end, cells were starved for 48 h and subsequently labeled with [32P]-orthophosphate for 1 h at different times after protein synthesis reactivation by addition of FBS as described in Materials and Methodology, and separated in 15% SDS-PAGE gel. The starvation step in this system was introduced to avoid incorporation of [32P]-orthophosphate in the peptide backbone and to look for novel protein phosphorylation. The result showed a phosphorylated E7 protein with an apparent molecular weight of 17.5 kDa (E7a1) that accumulates over the time, however, the 16 kDa protein band (E7b) was not observed at any time, suggesting that this E7 isoform is not phosphorylated (Fig. 4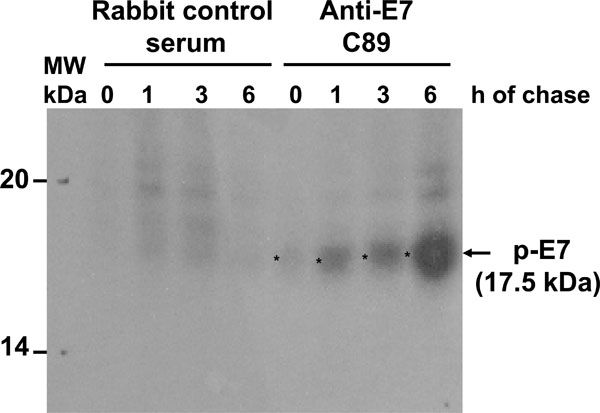 ). Several reports have shown that E7 can be phosphorylated in at least 2 different sites [21Massimi P, Pim D, Kühne C, Banks L. Regulation of the human papillomavirus oncoproteins by differential phosphorylation Mol Cell Biochem 2001; 227: 137-44., 30Smotkin D, Wettstein FO. The major human papillomavirus protein in cervical cancers is a cytoplasmic phosphoprotein J Virol 1987; 61: 1686-9., 31Storey A, Almond N, Osborn K, Crawford L. Mutations in the human papillomavirus type 16 E7 gene that affect transformation, transactivation and phosphorylation by E7 protein J Gen Virol 1990; 71: 965-70.], and this could explain the presence of a broad band that could contain the 17.5 kDa (E7a1) and the 17 kDa (E7a) proteins that were observed in the 2D gels.
). Several reports have shown that E7 can be phosphorylated in at least 2 different sites [21Massimi P, Pim D, Kühne C, Banks L. Regulation of the human papillomavirus oncoproteins by differential phosphorylation Mol Cell Biochem 2001; 227: 137-44., 30Smotkin D, Wettstein FO. The major human papillomavirus protein in cervical cancers is a cytoplasmic phosphoprotein J Virol 1987; 61: 1686-9., 31Storey A, Almond N, Osborn K, Crawford L. Mutations in the human papillomavirus type 16 E7 gene that affect transformation, transactivation and phosphorylation by E7 protein J Gen Virol 1990; 71: 965-70.], and this could explain the presence of a broad band that could contain the 17.5 kDa (E7a1) and the 17 kDa (E7a) proteins that were observed in the 2D gels.
Cellular localization of the different forms of HPV-16 E7 proteins.
Up to this point, the results showed that HPV-16 E7 from CaSki cells is processed in 3 different molecular weight forms, one of which is phosphorylated (E7a1). We also demonstrated that polyclonal and mAbs recognized different forms of the E7 protein. Taken together, this information made us wonder about the cellular localization of the 3 isoforms of E7. Thus, we used the different polyclonal and mAbs in cells fixed for immunofluorescence and co-localized the E7 protein with antibodies against cellular markers for nucleus (p21), endoplasmic reticulum (ER; calnexin) and Golgi (GM130), as previously reported [32Tominaga K, Kondo C, Kagata T, Hishida T, Nishizuka M, Imagawa M. The novel gene fad158, having a transmembrane domain and leucine-rich repeat, stimulates adipocyte differentiation J Biol Chem 2004; 279: 34840-8.-34Perez F, Pernet-Gallay K, Nizak C, Goodson HV, Kreis TE, Goud B. CLIPR-59, a new trans-Golgi/TGN cytoplasmic linker protein belonging to the CLIP-170 family J Cell Biol 2002; 156: 631-42.]. The results in Fig. (5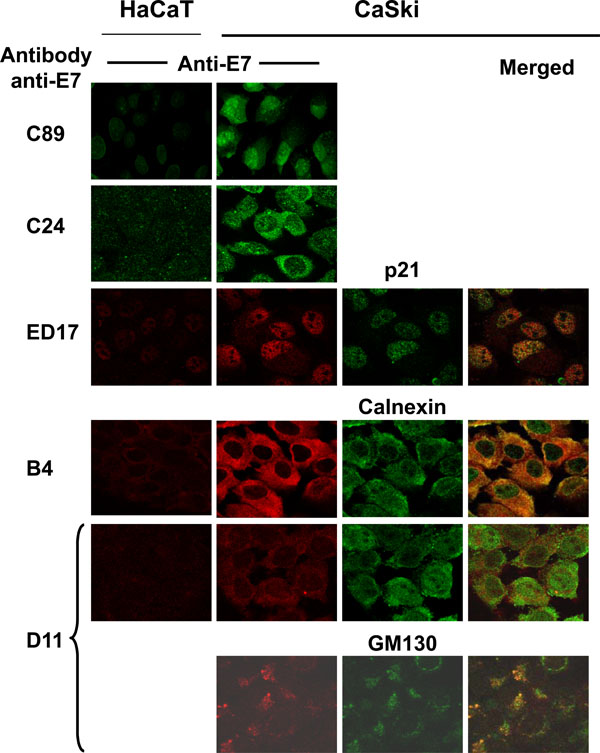 ) showed that the polyclonal antibody C89 recognized the E7 protein in the nucleus as wells as in cytoplasm, while the C24 polyclonal mainly stained the cytoplasm. When mAbs were tested together with the cellular markers it was evident that the ED17 antibody recognized the E7 protein in the nuclear compartment as this antibody co-localized only with the p21 nuclear marker (Fig. 5
) showed that the polyclonal antibody C89 recognized the E7 protein in the nucleus as wells as in cytoplasm, while the C24 polyclonal mainly stained the cytoplasm. When mAbs were tested together with the cellular markers it was evident that the ED17 antibody recognized the E7 protein in the nuclear compartment as this antibody co-localized only with the p21 nuclear marker (Fig. 5 , ED17, merged). The E7 protein recognized by the B4 mAb co-localized only with the calnexin marker, which suggests that the E7 form recognized by this antibody is present in the ER (Fig. 5
, ED17, merged). The E7 protein recognized by the B4 mAb co-localized only with the calnexin marker, which suggests that the E7 form recognized by this antibody is present in the ER (Fig. 5 , B4, merged). However, when the D11 mAb was tested a small part of this antibody co-localized with the calnexin marker of ER and a stronger co-localization signal was observed with the Golgi GM130 marker (Fig. 5
, B4, merged). However, when the D11 mAb was tested a small part of this antibody co-localized with the calnexin marker of ER and a stronger co-localization signal was observed with the Golgi GM130 marker (Fig. 5 , D11, merged). These results demonstrated that the E7 protein is recognized in different cellular compartments by different antibodies.
, D11, merged). These results demonstrated that the E7 protein is recognized in different cellular compartments by different antibodies.
In Vivo Cellular Processing of HPV-16 E7 Protein in Transfected Cells.
The presence of different forms of E7 localized in different cellular compartments prompted us to analyze the processing of the E7 protein in the cell. To analyze the E7 processing, Cos-7 cells were transiently transfected with the pcDNA-E7 plasmid to investigate de novo expression and localization of E7 protein at different periods of time and tested for immunofluorescence, as described in Materials and Methodology. The results in Fig. (6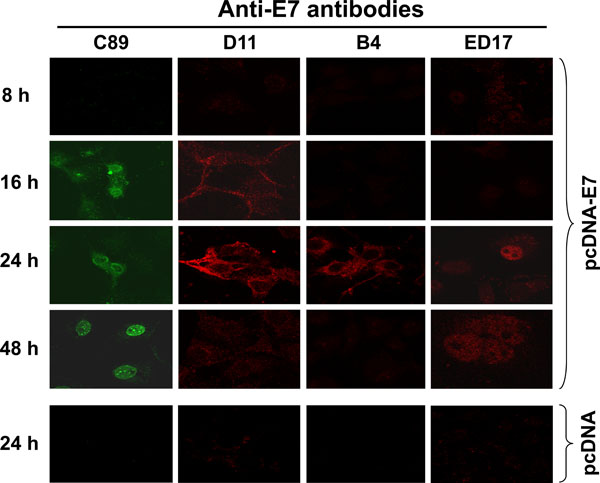 ) show that the polyclonal C89 antibody identified 3 different patterns of E7 staining; at first E7 is recognized by 16 h after transfection in the cytoplasm of the cells. By 24 h the E7 protein became concentrated in the periphery of the nucleus and by 48 h the fluorescence was only observed into the nucleus and concentrated in the nucleolus (Fig. 6
) show that the polyclonal C89 antibody identified 3 different patterns of E7 staining; at first E7 is recognized by 16 h after transfection in the cytoplasm of the cells. By 24 h the E7 protein became concentrated in the periphery of the nucleus and by 48 h the fluorescence was only observed into the nucleus and concentrated in the nucleolus (Fig. 6 , C89). With the D11 antibody (identifies E7 in ER and Golgi) a very light cytoplasmic fluorescence signal was observed by 16 h after E7 transfection, that became very strong by 24 h (Fig. 6
, C89). With the D11 antibody (identifies E7 in ER and Golgi) a very light cytoplasmic fluorescence signal was observed by 16 h after E7 transfection, that became very strong by 24 h (Fig. 6 , D11). However, the fluorescence signal disappeared totally by 48 h after Cos-7 cells transfection. In contrast, the B4 mAb that recognized only the E7 protein in the ER showed a strong fluorescence signal only at 24 h after E7 transfection (Fig. 6
, D11). However, the fluorescence signal disappeared totally by 48 h after Cos-7 cells transfection. In contrast, the B4 mAb that recognized only the E7 protein in the ER showed a strong fluorescence signal only at 24 h after E7 transfection (Fig. 6 , B4). While the ED17 mAb that recognized E7 protein only in the nucleus identified the protein only after 24 h and the fluorescence signal was still present by 48 h (Fig. 6
, B4). While the ED17 mAb that recognized E7 protein only in the nucleus identified the protein only after 24 h and the fluorescence signal was still present by 48 h (Fig. 6 , ED17). In these experiments the presence of E7 expression was not detected, with any of the different antibodies tested, before the 16 h and after 48 h (data not shown). The specificity of each one of the antibodies was determined by immunofluorescence using Cos-7 transfected cells with pcDNA plasmid alone as it was done for the pcDNA-E7 plasmid (data not shown). In Fig. (6
, ED17). In these experiments the presence of E7 expression was not detected, with any of the different antibodies tested, before the 16 h and after 48 h (data not shown). The specificity of each one of the antibodies was determined by immunofluorescence using Cos-7 transfected cells with pcDNA plasmid alone as it was done for the pcDNA-E7 plasmid (data not shown). In Fig. (6 ), only the 24 h time pcDNA control is shown for each one of the antibodies as this was the time of the highest E7 expression in the transfected cells.
), only the 24 h time pcDNA control is shown for each one of the antibodies as this was the time of the highest E7 expression in the transfected cells.
DISCUSSION
The E7 protein is a very well studied protein that had been demonstrated to interact with a variety of target proteins such as the pocket proteins (pRb, p130 and p107) [14Caldeira S, Dong W, Tommasino M. Analysis of E7/Rb associations Methods Mol Med 2005; 119: 363-79., 15Collins AS, Nakahara T, Do A, Lambert PF. Interactions with pocket proteins contribute to the role of human papillomavirus type 16 E7 in the papillomavirus life cycle J Virol 2005; 79: 14769-80.], proteins involved in cell cycle (E2F-Cyclin A complex, Cyclin E, p21Waf1, p27Kip1) [35Arroyo M, Bagchi S, Raychaudhuri P. Association of the human papillomavirus type 16 E7 protein with the S-phase-specific E2F-cyclin A complex Mol Cell Biol 1993; 13: 6537-46.-39Zerfass-Thome K, Zwerschke W, Mannhardt B, Tindle R, Botz JW, Jansen-Durr P. Inactivation of the cdk inhibitor p27KIP1 by the human papillomavirus type 16 E7 oncoprotein Oncogene 1996; 13: 2323-30.], transcription factors (pCAF acetyltransferase, TBP, AP-1 family factors) [40Maldonado E, Cabrejos ME, Banks L, Allende JE. Human papillomavirus-16 E7 protein inhibits the DNA interaction of the TATA binding transcription factor J Cell Biochem 2002; 85: 663-9.-43Antinore MJ, Birrer JM, Patel D, Nader L, McCance DJ. The human papillomavirus type 16 E7 gene product interacts with and trans-actives the AP1 family of transcription factors EMBO J 1996; 15: 1950-60.] and with the senescence-regulating protein DEK [44Wise-Draper TM, Allen HV, Thobe MN, et al. The human DEK proto-oncogene is a senescence inhibitor and an upregulated target of high-risk human papillomavirus E7 J Virol 2005; 79: 14309-17.], among others. All of these target proteins are present in the nucleus; however the reports about the distribution of the E7 in the cell have shown that this protein is present not only in nucleus but also in the cytoplasm. We believe that this differential distribution of E7 in the cells is due to the presence of different isoforms of E7 that are produced during its cellular processing. To test this hypothesis, monoclonal and polyclonal antibodies that recognize different epitopes in the E7 protein were developed in our laboratory. The results showed the presence of 3 isoforms of HPV-16 E7 protein that vary in molecular weight and IEP. These E7 proteins were described as E7a1 (17.5 kDa and IEP of 4.68), E7a (17 kDa and IEP of 6.18) and E7b (16 kDa and IEP of 6.96). The half-lives of the E7 proteins were calculated as 50 min for E7a and 70 min for E7b. These results are consistent with a previous report that showed that the half-life of E7 from CaSki cells was 55 min and 70 min in SiHa cells, although these researchers only identified one E7 band protein in each cell line [30Smotkin D, Wettstein FO. The major human papillomavirus protein in cervical cancers is a cytoplasmic phosphoprotein J Virol 1987; 61: 1686-9.]. The difference between Smotkin and Wettstein results and ours was the use of different anti-E7 antibodies and that these recognized the different isoforms of the E7 protein.
In the pulse-chase experiments we identified that E7 is initially synthesized as a 17 kDa protein and after approximately 1 h, it is processed to a faster moving band of 16 kDa with a short half-life. These results suggest that E7 undergoes some posttranslational modifications that generate the shift in molecular weight. A known modification for E7 is the phosphorylation of the serine residues 31 and 32 [29Barbosa MS, Edmonds C, Fisher C, Schiller JT, Lowy DR, Vousden KH. The region of the HPV E7 oncoprotein homologous to adenovirus E1a and SV40 large T antigen contains separate domains for Rb binding and casein kinase II phosphorylation EMBO J 1990; 9: 153-60., 45Firzlaff JM, Lüscher B, Eisenman RN. Negative charge at the casein kinase II phosphorylation site is important for transformation but not for Rb protein binding by the E7 protein of human papillomavirus type 16 Proc Natl Acad Sci USA 1991; 88: 5187-91.], although serine 71 is also a potential phosphorylation site [31Storey A, Almond N, Osborn K, Crawford L. Mutations in the human papillomavirus type 16 E7 gene that affect transformation, transactivation and phosphorylation by E7 protein J Gen Virol 1990; 71: 965-70., 46Massimi P, Banks L. Differential phosphorylation of the HPV-16 E7 oncoprotein during the cell cycle Virology 2000; 276: 388-94.]. Our results showed a broad phosphorylated band of a calculated molecular weight of 17.5 kDa that could be the 17 kDa band that was recognized in the immunoprecipitations. The difference in molecular weight could be due to the change in charge that makes the protein to be retarded in the SDS-PAGE gel. Another possibility is that the antibodies did not recognize efficiently the non-phosphorylated protein as has been reported for other mAbs [47Kee SH, Choi YO, Song YS, Lee HP, Chang WH. Identification of antigenic differences between the phosphorylated and nonphosphorylated forms of the E7 protein of human papillomavirus type 16 J Med Virol 1998; 54: 129-34.], suggesting that the phosphorylated E7 protein has distinct antigenic properties (conformational changes). However, these results did not account for the lower molecular weight band of 16 kDa (E7b) that was identified in the Western blot and in the immunoprecipitations. Another as yet unidentified modification could exist and be the reason for the presence of this faster moving E7b protein.
According to the HPV-16 E7 protein sequence the calculated IEP of this protein is 4.05, however, when this characteristic was measured with recombinant E7 protein produced in bacteria the IEP obtained was 5.4 [48Pahel G, Aulabaugh A, Short SA, et al. Structural and functional characterization of the HPV16 E7 protein expressed in bacteria J Biol Chem 1993; 268: 26018-25.]. When the IEP of the HPV-18 E7 protein produced in vitro (using rabbit reticulocyte lysates) or immunoprecipitated from HeLa cells, there were 3 bands of the same molecular weight, but different IEP [49Roggenbuck B, Larsen PM, Fey SJ, Bartsch D, Gissmann L, Schwarz E. Human papillomavirus type 18 E6*, E6 and E7 protein synthesis in cell-free translation systems and comparison of E6 and E7 in vitro translation products to proteins immunoprecipitated from human epithelial cells J Virol 1991; 65: 5068-72.]. The researchers suggested that this was a typical pattern for phosphorylated proteins and this was similar to what we found. However, in our experiments not only the IEP, but also the molecular weights of the E7 proteins (E7a1, E7a and E7b), were different in the 2D gels. This again suggests that the C89 polyclonal antibody is recognizing different epitopes of the E7 isoforms that have not been fully characterized.
At least 2 distinct molecular weights have been reported for HPV-16 E7 by 2 different groups [50Tommasino M, Contorni M, Scarlato V, Bugnoli M, Maundrell K, Cavalieri F. Synthesis, phosphorylation, and nuclear localization of human papillomavirus E7 protein in Schizosaccharomyces pombe Gene 1990; 93: 265-70., 51Kanda T, Zanma S, Watanabe S, Furuno A, Yoshiike K. Two Inmunodominant regions of the human papillomavirus type 16 E7 protein are masked in the nuclei of monkey COS-1 cells Virology 1991; 182: 723-31.]. However, the C89 antibody was able to recognize simultaneously 2 forms of E7 by radio-immunoprecipitation and 3 when 2D gels were used. It has been shown that E7 is highly hydrophobic and that this characteristic together with its conformational structure produce an anomalous electrophoretic behavior. Purified E7 protein is present as oligomers, and tends to be soluble and with a molecular weight close to the calculated in the presence of 8M urea [13Alonso LG, García-Alai MM, Nadra AD, et al. High-Risk (HPV16) Human Papillomavirus E7 oncoprotein is highly stable and extended, with conformational transitions that could explain its multiple cellular binding partners Biochemistry 2002; 41: 10510-8., 52Alonso LG, Garcia-Alai MM, Smal C, et al. The HPV16 E7 viral oncoprotein self-assembles into defined spherical oligomers Biochemistry 2004; 43: 3310-7.]. It is possible that we were able to visualize the 3 forms of E7 in the 2D gels, not only because the difference in charge, but because the proteins were solubilized in 8M urea.
When we examined the localization of the different E7 forms in the cellular compartments by immunofluorescence staining, it was observed that the E7 protein was first observed in the ER as this was the first signal observed with the antibodies (16 h). Subsequently, the E7 protein was visualized in the Golgi compartment (16 to 24 h) and finally translocated to the nucleus (48 h), where it probably interacts with target proteins, such as pRb. From this work, it is clear that the E7b (16 KDa, recognized by mAbs) isoform is localized in the cytoplasm and nucleus, but the localization of the 17 and 17.5 kDa E7 isoforms still needs to be determined as there are not specific antibodies available to differentiate them. According with this processing of the E7 protein, it is possible that the protein encounters some posttranslational modifications during the transit through different cellular compartments that allow the final localization of the protein. Studying the amino acid sequence of the HPV-16 E7 protein, it could be observed that it contains a consensus sequence for glycosylation of Asp 29, has 2 sulfation sites (Tyr 23 and 26) and 5 phosphorylation sites besides the already known Ser 31, 32 and 71 [46Massimi P, Banks L. Differential phosphorylation of the HPV-16 E7 oncoprotein during the cell cycle Virology 2000; 276: 388-94.]. Up today, only phosphorylation has been reported for the E7 protein and this posttranslational modification only accounts for the difference in charge identified in the 3 different forms of E7, but other modifications will still need to be studied to clarify the differences on molecular weight identified by our polyclonal and mAbs.
During the processing of the E7 protein in the Cos-7 transfected cells, the protein is localized at the perinuclear zone by 24 h after being synthesized. This localization is in agreement with a previous report that showed that E7 from HPV-16 was observed in ER, cytoplasm and the nuclear membrane, where E7 was able to activate the α-glucosidase enzyme in an allosteric way [18Zwerschke W, Mannhardt B, Massimi P, et al. Allosteric activation of acid α-glucosidase by the human papillomavirus E7 protein J Biol Chem 2000; 275: 9534-41.]. Subsequently, during the expression of the E7 protein in the Cos-7 transfected cells (48 h), the protein was visualized in the nucleus and relocalized into the nucleolus of the cells. These structures have also been reported to be present in CaSki cells, but only during the G2/M cell cycle phase [53Zatsepina O, Braspenning J, Robberson D, et al. The human papillomavirus type 16 E7 protein is associated with the nucleolus in mammalian and yeast cells Oncogene 1997; 14: 1137-45.]. The nuclear localization of the E7 protein has been reported in several studies [50Tommasino M, Contorni M, Scarlato V, Bugnoli M, Maundrell K, Cavalieri F. Synthesis, phosphorylation, and nuclear localization of human papillomavirus E7 protein in Schizosaccharomyces pombe Gene 1990; 93: 265-70., 51Kanda T, Zanma S, Watanabe S, Furuno A, Yoshiike K. Two Inmunodominant regions of the human papillomavirus type 16 E7 protein are masked in the nuclei of monkey COS-1 cells Virology 1991; 182: 723-31., 53Zatsepina O, Braspenning J, Robberson D, et al. The human papillomavirus type 16 E7 protein is associated with the nucleolus in mammalian and yeast cells Oncogene 1997; 14: 1137-45., 54Angeline M, Merle E, Moroianu J. The E7 oncoprotein of high-risk human papillomavirus type 16 enters the nucleus via a nonclasical Ran-dependent pathway Virology 2003; 317: 13-23.]; however, this protein does not contain a consensus sequence to be translocated into the nucleus. The translocation of the E7 protein into this compartment has been reported to be through a non-conventional Ran-GTP pathway and an intermediary carrier protein seems to be involved [54Angeline M, Merle E, Moroianu J. The E7 oncoprotein of high-risk human papillomavirus type 16 enters the nucleus via a nonclasical Ran-dependent pathway Virology 2003; 317: 13-23.]. Alternatively, in vitro characterization of E7 protein has demonstrated that formation of dimers and tetramers of E7 depends on pH changes which allow exposure of hydrophobic epitopes [13Alonso LG, García-Alai MM, Nadra AD, et al. High-Risk (HPV16) Human Papillomavirus E7 oncoprotein is highly stable and extended, with conformational transitions that could explain its multiple cellular binding partners Biochemistry 2002; 41: 10510-8.]. It is also well established that cell compartments are microenvironments that functions at different pHs [55Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase Microbiol Mol Biol Rev 2006; 70: 177-91., 56Wiseman RL, Koulov A, Powers E, Kelly JW, Balch WE. Protein energetics in maturation of the early secretory pathway Curr Opin Cell Biol 2007; 19: 359-67.]. It is possible then, that final localization of the different isoforms of E7 in the cell could be drived by exposure of different epitopes due to oligomerization of E7 induced by changes in pH, during their transit through the different cellular compartments.
The low levels of E7 expression of the high risk HPVs, together with the short half-life of this protein, makes it difficult to carry on with biochemical experiments helpful to completely characterize this protein. Most of the information obtained in an effort to biochemically characterize E7 protein has been generated from recombinant E7 protein produced in bacteria or produced under in vitro conditions. However, some of the posttranslational modifications of the proteins are predominantly observed in eukaryotic cells. The lack of these modifications would change the molecular weights as well as the net charges of the protein, making it difficult to detect the intermediates or processed forms of the HPV-16 E7 protein that are reported in this paper as we used eukaryotic cell cultures. In this way, 3 forms of HPV-16 E7 proteins were identified with polyclonal and mAbs that showed different molecular weights and different IEPs (E7a1, E7a and E7b). Due to the fact that the different antibodies recognized different epitopes, the E7 protein was recognized in different cellular compartments at different times during its processing.
The E7 protein has been recognized as a multifunctional protein that interacts with a high variety of target proteins. It will be of great interest to identify the complete molecular processing of the E7 protein (other posttranslational modifications) that will allow a better knowledge about the structure and the biological activity of this oncogenic protein and its role in the transformation process.
ACKNOWLEDGEMENTS
We thank the technical support of the people from the Virus and Cancer Department from the National Institute of Public Health, Mexico. We also acknowledge to Dr. Robert Tindle and Dr. Eric Lam for helpful comments and to the MSc Tanya Plett for revising the manuscript.
Potential Conflicts of Interest
The funds for this work were obtained from the Instituto Nacional de Salud Publica and supported by the Mexican government through a grant from CONACYT-SEP (49574-24483-M).
H. Valdovinos-Torres, is PhD student of the Postgraduate Program of Ciencias Biológicas, UNAM, Mexico City, Mexico. The PhD supervisor of this student was Dr. Lourdes Gutierrez-Xicotencatl.








