- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
The Role of Microtubule Association in Plasmodesmal Targeting of Potato mop-top virus Movement Protein TGBp1
Elena A Shemyakina1, Andrey G Solovyev2, 3, Olga G Leonova4, Vladimir I Popenko4, Joachim Schiemann5, Sergey Yu Morozov*, 1, 2
Abstract
Cell-to-cell movement of Potato mop-top virus (PMTV) is mediated by three virus-encoded ‘triple gene block’ (TGB) proteins termed TGBp1, TGBp2 and TGBp3. TGBp1 binds virus RNAs to form viral ribonucleoprotein complexes (vRNPs), the transport form of viral genome. TGBp2 and TGBp3 are necessary for intracellular delivery of TGBp1-containing vRNPs to plasmodesmata. To analyze subcellular localization and transport of TGBp1 we used a single binary vector for agrobacterium-mediated co-expression of PMTV TGBp1 fused to green fluorescent protein and TGBp2/TGBp3. At two days post infiltration (dpi) TGBp1 was found in the nucleus and in association with microtubules (MTs). Similar localization pattern was revealed in cells expressing GFP-TGBp1 alone after particle bombardment. At 3 dpi, in addition to the nucleus and MTs, TGBp1 was detected in numerous granular bodies located both along the MTs and at the cell wall. The latter structures co-localized with plasmodesmata-associated callose depositions. At 4 dpi, GFP-TGBp1 was detected in cell wall-associated bodies and also in residual MTs, the nucleoplasm and large perinuclear inclusions resembling aggresomes. Therefore GFP-TGBp1 association with MTs preceded to its localization to plasmodesmata. Disassembly of cell MTs by colchicine prevented GFP-TGBp1 targeting to plasmodesmata and the MT-dependent aggresome formation. Deletion analysis also revealed a correlation between TGBp1 microtubule association and plasmodesmata targeting. We propose that TGBp1 interaction with MTs may be important for the formation of vRNP bodies destined for the transport to plasmodesmata as well as degradation of the excessive TGBp1.
Article Information
Identifiers and Pagination:
Year: 2011Volume: 5
First Page: 1
Last Page: 11
Publisher Id: TOVJ-5-1
DOI: 10.2174/1874357901105010001
Article History:
Received Date: 3/8/2010Revision Received Date: 11/10/2010
Acceptance Date: 25/10/2010
Electronic publication date: 5/1/2011
Collection year: 2011
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http: //creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the A. N. Belozersky Institute of Physico-Chemical Biology, Moscow State University, 119992 Moscow, Russia; Tel: +7 495 939 31 98; Fax: +7 495 939 31 81; E-mail: morozov@genebee.msu.su
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 3-8-2010 |
Original Manuscript | The Role of Microtubule Association in Plasmodesmal Targeting of Potato mop-top virus Movement Protein TGBp1 | |
INTRODUCTION
Cell-to-cell transport of plant viruses occurs through plasmodesmata, the channels interconnecting neighboring cells in plant tissues, and requires dedicated virus-encoded movement proteins (MPs) [1Lucas WJ. Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes Virology 2006; 344: 169-84.-5Lucas WJ, Ham BK, Kim JY. Plasmodesmata - bridging the gap between neighboring plant cells Trends Cell Biol 2009; 19: 495-503.]. In positive-stranded RNA viruses the number of encoded MPs can vary between distant virus groups, and they are often unrelated in their primary structure [1Lucas WJ. Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes Virology 2006; 344: 169-84.]. Nevertheless, viral MPs which represent dissimilar transport systems share three characteristic features. First, the RNA-binding activity is believed to be responsible for formation of complexes with viral genomic RNAs (viral ribonucleoproteins, vRNPs) which represent the transport form of viral genomes capable of cell-to-cell movement. Second, the ability of MPs to interact with cell membranes, in particular with the endoplasmic reticulum (ER), may direct MPs to specific pathway of intracellular transport to plasmodesmata. Third, MPs are able to accumulate in plasmodesmata or plasmodesmata-associated sites and to increase the size exclusion limit of plasmodesmata pores for efficient passage of macromolecules to neighboring cells [3Taliansky M, Torrance L, Kalinina NO. Role of plant virus movement proteins Methods Mol Biol 2008; 451: 33-54.-5Lucas WJ, Ham BK, Kim JY. Plasmodesmata - bridging the gap between neighboring plant cells Trends Cell Biol 2009; 19: 495-503.].
A number of plant virus genera require for their cell-to-cell movement a ‘triple gene block’ (TGB), conserved gene module encoding three MPs [6Morozov SY, Dolja VV, Atabekov JG. Probable reassortment of genomic elements among elongated RNA-containing plant viruses J Mol Evol 1989; 29: 52-62.]. The TGB position relative to other genes vary in genomes of different viruses, however the order of genes within the TGB and the properties of the encoded proteins are conserved [7Morozov SY, Solovyev AG. Triple gene block: modular design of a multifunctional machine for plant virus movement J Gen Virol 2003; 84: 1351-66., 8Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. Varied movement strategies employed by triple gene block-encoding viruses Mol Plant-Microbe Interact 2010; 23: 1231-47.]. Based on differences in the structure of TGB proteins and the requirement of viral capsid protein for cell-to-cell movement two types of TGB are distinguished: ‘hordei-like’ TGB in rod-shaped viruses (family Virgaviridae and genus Benyvirus) and ‘potex-like’ TGB in filamentous viruses (family Flexiviridae) [7Morozov SY, Solovyev AG. Triple gene block: modular design of a multifunctional machine for plant virus movement J Gen Virol 2003; 84: 1351-66., 8Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. Varied movement strategies employed by triple gene block-encoding viruses Mol Plant-Microbe Interact 2010; 23: 1231-47.].
Studies of hordei-like TGB carried out in the recent years and employed as models Barley stripe mosaic virus (BSMV, genus Hordeivirus), Poa semilatent virus (genus Hordeivirus) and Potato mop-top virus (PMTV, genus Pomovirus) revealed characteristic biochemical properties and functions of individual TGB proteins, as well as their physical and functional interactions involved in virus cell-to-cell movement [8Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. Varied movement strategies employed by triple gene block-encoding viruses Mol Plant-Microbe Interact 2010; 23: 1231-47., 9Jackson AO, Lim HS, Bragg J, Ganesan U, Lee MY. Hordeivirus replication, movement, and pathogenesis Annu Rev Phytopathol 2009; 47: 385-422.]. TGBp1, the largest of TGB proteins with molecular mass of 51 to 63 kDa in hordei-like TGBs, consists of three domains, the N-terminal domain, internal domain and the C-terminal NTPase/helicase domain [10Makarov VV, Rybakova EN, Efimov AE, et al. Domain organization of the N-terminal portion of hordeivirus movement protein TGBp1 J Gen Virol 2009; 90: 3022-2.]. In Poa semilatent virus, the N-terminal domain represents an unfolded protein region which includes two positively charged sequence motifs involved in virus long-distance movement via the phloem, whereas the internal domain is a small structured domain able to interact with both the N-terminal domain and the NTPase/helicase domain [10Makarov VV, Rybakova EN, Efimov AE, et al. Domain organization of the N-terminal portion of hordeivirus movement protein TGBp1 J Gen Virol 2009; 90: 3022-2., 11Kalinina NO, Rakitina DA, Yelina NE, et al. RNA-binding properties of the 63 kDa protein encoded by the triple gene block of poa semilatent hordeivirus J Gen Virol 2001; 82: 2569-78.]. The latter domain has the NTPase and helicase activities and is capable of self-interactions [10Makarov VV, Rybakova EN, Efimov AE, et al. Domain organization of the N-terminal portion of hordeivirus movement protein TGBp1 J Gen Virol 2009; 90: 3022-2.-14Leshchiner AD, Solovyev AG, Morozov SY, Kalinina NO. A minimal region in the NTPase/helicase domain of the TGBp1 plant virus movement protien is responsible for ATPase activity and cooperative RNA binding J Gen Virol 2006; 87: 3087-95.]. Additionally, all three domains of the hordeiviral TGBp1 bind RNA exhibiting either a cooperative (the internal and the NTPase/helicase domains) or non-cooperative (the N-terminal domain) binding [10Makarov VV, Rybakova EN, Efimov AE, et al. Domain organization of the N-terminal portion of hordeivirus movement protein TGBp1 J Gen Virol 2009; 90: 3022-2., 11Kalinina NO, Rakitina DA, Yelina NE, et al. RNA-binding properties of the 63 kDa protein encoded by the triple gene block of poa semilatent hordeivirus J Gen Virol 2001; 82: 2569-78., 13Donald RG, Lawrence DM, Jackson AO. The barley stripe mosaic virus 58-kilodalton beta(b) protein is a multifunctional RNA binding protein J Virol 1997; 71: 1538-46.]. These RNA binding activities are responsible for interaction with viral RNAs and the formation of vRNPs; such RNPs were isolated from BSMV-infected plants and were shown to contain BSMV positive-stranded RNAs and TGBp1 but not other viral or non-viral components [15Lim H-S, Bragg JN, Ganesan U, et al. Triple gene block protein interactions involved in movement of Barley stripe mosaic virus J Virol 2008; 82: 4991-5006.]. The presence of multiple binding sites in TGBp1 and the ability of internal domain for interactions with other protein domains, as well as the proposed ability of the N-terminal domain to conditionally adopt a structured conformation, provide the basis for structural remodeling of vRNPs that would be necessary for different stages of virus movement in plant [10Makarov VV, Rybakova EN, Efimov AE, et al. Domain organization of the N-terminal portion of hordeivirus movement protein TGBp1 J Gen Virol 2009; 90: 3022-2.]. TGBp2 and TGBp3 are small integral membrane proteins required for delivery of hordei-like TGBp1 to plasmodesmata [7Morozov SY, Solovyev AG. Triple gene block: modular design of a multifunctional machine for plant virus movement J Gen Virol 2003; 84: 1351-66., 8Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. Varied movement strategies employed by triple gene block-encoding viruses Mol Plant-Microbe Interact 2010; 23: 1231-47.]. While TGBp2, when expressed in the absence of TGBp3, is localized to the cortical endoplasmic reticulum (ER) or numerous ER-associated vesicular bodies, TGBp3 localizes to plasmodesmata or plasmodesmata-associated membrane compartments called peripheral membrane bodies (PMBs) and, moreover, is able to target TGBp2 from the ER structures to plasmodesmata and PMBs [7Morozov SY, Solovyev AG. Triple gene block: modular design of a multifunctional machine for plant virus movement J Gen Virol 2003; 84: 1351-66.-9Jackson AO, Lim HS, Bragg J, Ganesan U, Lee MY. Hordeivirus replication, movement, and pathogenesis Annu Rev Phytopathol 2009; 47: 385-422.]. Furthermore, TGBp2 and TGBp3 direct TGBp1 to PMBs and plasmodesmata, and in the case of BSMV this targeting could involve direct interactions between TGBp1 and TGBp3 [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30., 17Lim H-S, Bragg JN, Ganesan U, et al. Subcellular localization of the barley stripe mosaic virus triple gene block proteins J Virol 2009; 83: 9432-48.]. Recent evidences suggest that TGBp3 contains signals of plasmodesmata targeting [18Schepetilnikov MV, Solovyev AG, Gorshkova EN, et al. Intracellular targeting of a hordeiviral membrane-spanning movement protein: sequence requirements and involvement of an unconventional mechanism J Virol 2008; 82: 1284-93., 19Tilsner J, Cowan GH, Roberts AG, et al. Plasmodesmal targeting and intercellular movement of potato mop-top pomovirus is mediated by a membrane anchored tyrosine-based motif on the lumenal side of the endoplasmic reticulum and the C-terminal transmembrane domain in the TGB3 movement protein Virology 2010; 402: 41-51.] and that TGBp3 serves therefore as a ‘driving force’ for the targeting of TGBp2, TGBp1, and most probably TGBp1-containing vRNPs to plasmodesmata [8Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. Varied movement strategies employed by triple gene block-encoding viruses Mol Plant-Microbe Interact 2010; 23: 1231-47.].
Previously we analyzed PMTV TGB functions using an experimental system in which intra- and intercellular transport of TGB proteins was reconstructed using individually cloned PMTV TGB genes and a particle bombardment-based delivery of recombinant plasmids into plant cells [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.]. Specifically, this experimental system includes two expression vectors which carry, under the control of the cauliflower mosaic virus 35S promoter, either the PMTV TGBp1 gene fused to the green fluorescent protein (GFP) gene, or the cloned region of PMTV genome comprising overlapping genes of TGBp2 and TGBp3. The latter construct mimics the natural expression of TGBp2 and TGBp3 from a single subgenomic RNA [20Zhou H, Jackson AO. Expression of the barley stripe mosaic virus RNA beta ‘triple gene block’ Virology 1996; 216: 367-79.]. After bombardment of plant leaves with metal particles carrying the GFP-TGBp1 expression vector, the fusion protein expressed in individual epidermal cells was distributed uniformly in the cytoplasm and also found in the nucleus. However, when leaves were bombarded by a mixture of GFP-TGBp1 and TGBp2/TGBp3 expression vectors, GFP-TGBp1 was directed to large PMBs and to cell wall-embedded punctate bodies supposed to correspond to plasmodesmata and, moreover, was transported into cells immediately adjacent to the bombarded cell showing that PMTV TGBp2 and TGBp3 can mediate transport of GFP-TGBp1 to and through plasmodesmata in the absence of other virus-encoded components [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.]. Further experiments revealed that specific interactions between TGBp1 and TGBp2/TGBp3 are required for TGBp1 delivery to plasmodesmata and translocation to adjacent cells as well as allowed us to carry out a preliminary mapping of protein region involved in interaction between TGB proteins [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.]. Therefore, the experimental system based on co-expression of TGB genes by use of particle bombardment proved to be a useful tool for studies of TGB-mediated transport.
In this paper we report elaboration of an experimental system based on agrobacteria-mediated transient expression of three PMTV TGB proteins from a single binary vector providing controlled synthesis levels for TGBp1, TGBp2 and TGBp3. Using this system we demonstrate a transient association of GFP-tagged TGBp1 with microtubules and microtubule-associated bodies that preceded TGBp1 targeting to cell wall-embedded structures which are proved to be plasmodesmata. We also show that disruption of cell microtubules with colchicine prevents TGBp1 targeting to plasmodesmata, and TGBp1 mutations which block interactions with microtubules also impede its plasmodesmata targeting.
MATERIALS AND METHODOLOGY
Recombinant Clones
Recombinant plasmids pRT-GFP-TGBp1 [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.] and pCB-GFP-TAL [18Schepetilnikov MV, Solovyev AG, Gorshkova EN, et al. Intracellular targeting of a hordeiviral membrane-spanning movement protein: sequence requirements and involvement of an unconventional mechanism J Virol 2008; 82: 1284-93.] have been described earlier. To generate the YFP-TAL construct the GFP gene in pCB-GFP-TAL was replaced with the YFP gene using appropriate restriction sites. To obtain YFP-MAP4 fusion gene, the microtubule-binding domain of mouse microtubule-associated protein 4 (MAP4) was amplified with specific primers as described [21Marc J, Granger CL, Brincat J, et al. A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells Plant Cell 1998; 10: 1927-40.] and cloned as a fusion with the YFP gene into pRT100 [22Töpfer R, Matzeit V, Gronenborn B, Steinbiss H-H. A set of plant expression vectors for transcriptional and translational fusions Nucleic Acids Res 1987; 15: 5890.]. Expression cassettes including the 35S promoter, the fluorescent protein fusion gene and transcriptional terminator were cut out of the vectors pRT-GFP-TGBp1, pCB-YFP-TAL and pRT-YFP-MAP4 by digestion with HindIII and ligated into HindIII-digested binary vector pLH7000 [23Hausmann L, Töpfer R. Entwicklung von Plasmid-Vektoren Vortr Pflanzenzcht 1999; 45: 155-72. (in German)] provided by Dr L. Hausmann (Federal Centre for Breeding Research on Cultivated Plants, Germany). pLH-35S:GFP-TGBp1-NOS:TGBp2/TGBp3 was constructed in three steps. Using pBin19 [24Bevan M. Binary Agrobacterium vectors for plant transformation Nucleic Acids Res 1984; 12: 8711-21.] as a template, NOS promoter was amplified with primers 5’-GCGGGCCCGTCGACCTGTC AAACACTGATAGTTTA and 5’-GCGGATCCATGGTAC CTCGAGATCCGGTGCAGATTATTTG; NOS terminator was amplified with primers 5'-GCGGATCCTCTAGAGATC GTTCAAACATTTGGCAATAA and 5’-CGAGCTCGTCG ACCCGATCTAGTAACATAGATGA. The resulting products were digested with ApaI-BamHI and BamHI-SacI, respectively, and taken for a three-way ligation into ApaI-SacI-cut pGEM7zf+ (Promega). The resulting vector, which contained XhoI and XbaI restriction sites between the NOS promoter and terminator sequences, was digested with XhoI and XbaI and ligated with XhoI-XbaI-fragment of pRT-TGBp2/TGBp3 [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.] containing overlapping PMTV TGBp2 and TGBp3 genes. The expression cassette from the resulting vector was cut out with SalI and subcloned into SalI-digested pLH7000-GFP-TGBp1.
Particle Bombardment, Agroinfiltration and Fluorescent Microscopy
Particle bombardment of N. benthamiana leaves was performed using the flying disc method with a high-pressure helium-based PDS-1000 system (Bio-Rad) as described [25Morozov SY, Fedorkin ON, Juettner G, Schiemann J, Baulcombe D, Atabekov JG. Complementation of a potato virus X mutant mediated by bombardment of plant tissues with cloned viral movement protein genes J Gen Virol 1997; 78: 2077-83.]. Agroinfiltration and imaging of cells with a Leica TCS SP2 system was carried out as described [26Yelina NE, Erokhina TN, Lukhovitskaya NI, et al. Localization of Poa semilatent virus cysteine-rich protein in peroxisomes is dispensable for its ability to suppress RNA silencing J Gen Virol 2005; 86: 479-89.]. The software package provided by the manufacturer was used for projections of serial optical sections and image processing.
Leaf Treatments
Callose was visualized in leaf cells with 0.1% (w/v) sirofluor-containing aniline blue (Merck) in 1 M glycine, pH 9.5, as previously described [27Stone BA, Evans NA, Bonig I, Clarke AE. The application of sirofluor, a chemically defined fluorochrome from aniline blue for the histochemical detection of callose Protoplasma 1984; 122: 191-5.]. To induce plasmolysis, leaf pieces were incubated in 0.5 M mannitol solution for 5 - 10 min. The colchicine treatments were done by infiltrating 50 µM colchicine solution in an agroinfiltration buffer (10 mM MES [pH 5.85], 10 mM MgCl2, 150 µM acetosyringone) into N. benthamiana leaves.
RESULTS
TGBp1 Interaction with Microtubules in Particle Bombardment Assay
In our previous particle bombardment experiments, PMTV 35S:GFP-TGBp1 construct gave rise to diffuse fluorescence observed in the nucleus and the cytoplasm of bombarded cells. Importantly, the cytoplasmic TGBp1 fluorescence was not associated with the cell wall or plasmodesmata [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.]. Additionally, the cytoplasmic fluorescence was sometimes associated with numerous faint thin strings resembling in their appearance microtubules (MTs) (not published). In these earlier experiments, microscopy of bombarded leaves was carried out at 15 - 18 hours post bombardment that would hinder detection of the MT-like structures. Therefore in this study we used a longer incubation of bombarded leaves before microscopy. When GFP-TGBp1 was observed in epidermal cells of N. benthamiana leaves at 24 hours post bombardment or at later time points, strong association of GFP fluorescence with the MT-like structures was easily detected in most examined cells (Figs. 1A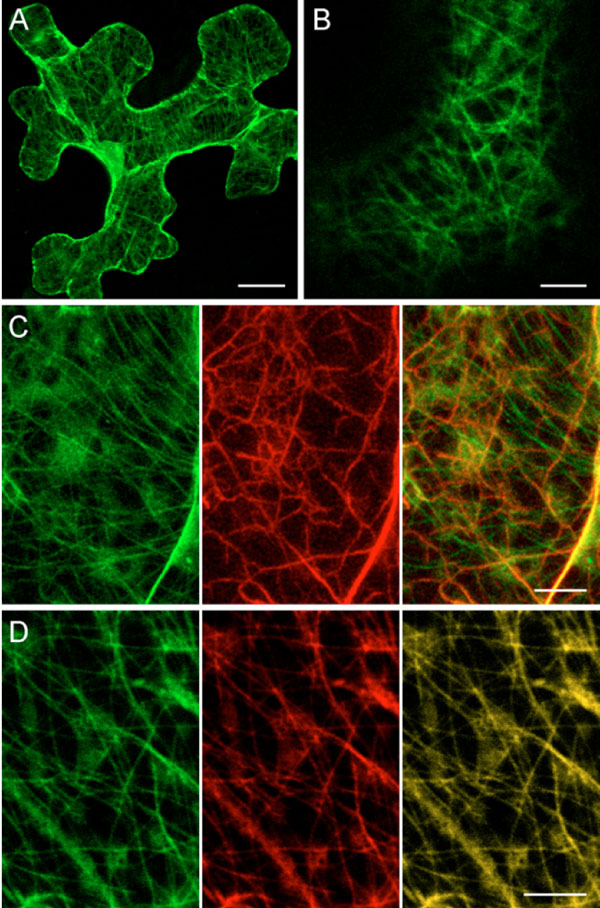 , 3B
, 3B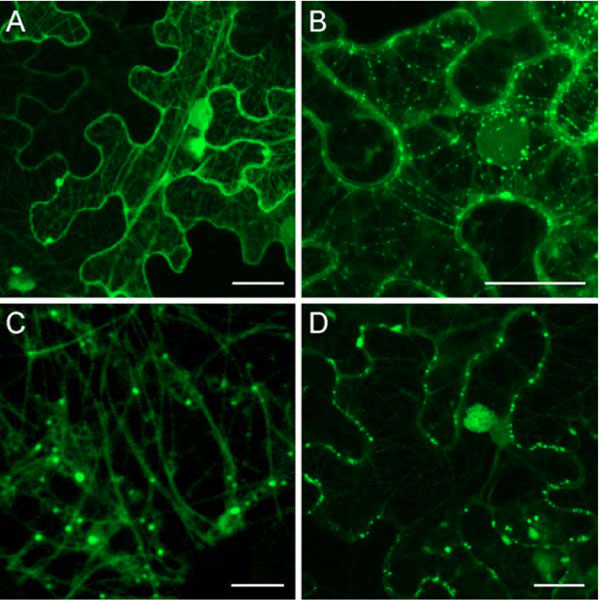 ). To identify the nature of the GFP-TGBp1-containing string-like structures, co-localization studies with markers of MTs and microfilaments were carried out. To visualize actin cables the actin-binding domain of mouse talin fused to yellow fluorescent protein (YFP) (YFP-Tal) known to bind the actin cytoskeleton in different types of living plant cells was used [28Kost B, Spielhofer P, Chua N-H. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes Plant J 1998; 16: 393-401.]. MTs were visualized by use of marker protein YFP-MAP4 that represented yellow fluorescent protein (YFP) fused to the microtubule-binding domain of mouse microtubule-associated protein 4 (MAP4) as described [21Marc J, Granger CL, Brincat J, et al. A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells Plant Cell 1998; 10: 1927-40.]. This fusion has been previously shown to bind MTs in plant cells [21Marc J, Granger CL, Brincat J, et al. A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells Plant Cell 1998; 10: 1927-40.]. Co-expression of GFP-TGBp1 with YFP-Tal by particle bombardment revealed that GFP and YFP fluorescent signals were associated with different non-overlapping filamentous structures (Fig. 1C
). To identify the nature of the GFP-TGBp1-containing string-like structures, co-localization studies with markers of MTs and microfilaments were carried out. To visualize actin cables the actin-binding domain of mouse talin fused to yellow fluorescent protein (YFP) (YFP-Tal) known to bind the actin cytoskeleton in different types of living plant cells was used [28Kost B, Spielhofer P, Chua N-H. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes Plant J 1998; 16: 393-401.]. MTs were visualized by use of marker protein YFP-MAP4 that represented yellow fluorescent protein (YFP) fused to the microtubule-binding domain of mouse microtubule-associated protein 4 (MAP4) as described [21Marc J, Granger CL, Brincat J, et al. A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells Plant Cell 1998; 10: 1927-40.]. This fusion has been previously shown to bind MTs in plant cells [21Marc J, Granger CL, Brincat J, et al. A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells Plant Cell 1998; 10: 1927-40.]. Co-expression of GFP-TGBp1 with YFP-Tal by particle bombardment revealed that GFP and YFP fluorescent signals were associated with different non-overlapping filamentous structures (Fig. 1C ). In contrast, in cells where GFP-TGBp1 and YFP-MAP4 were co-expressed the string-like structures containing GFP-TGBp1 coincided with MTs labeled with YFP-MAP4 (Fig. 1D
). In contrast, in cells where GFP-TGBp1 and YFP-MAP4 were co-expressed the string-like structures containing GFP-TGBp1 coincided with MTs labeled with YFP-MAP4 (Fig. 1D ). A similar co-localization with MAP4-labeled MTs was recently reported for Tobacco mosaic virus (TMV) MP [29Ouko MO, Sambade A, Brandner K, et al. Tobacco mutants with reduced microtubule dynamics are less susceptible to TMV Plant J 2010; 62: 829-39.]. Thus our observations demonstrated association of TGBp1 with MTs.
). A similar co-localization with MAP4-labeled MTs was recently reported for Tobacco mosaic virus (TMV) MP [29Ouko MO, Sambade A, Brandner K, et al. Tobacco mutants with reduced microtubule dynamics are less susceptible to TMV Plant J 2010; 62: 829-39.]. Thus our observations demonstrated association of TGBp1 with MTs.
Expression of Three PMTV TGB Proteins from a Single Binary Vector
Observations of GFP-TGBp1 in association with MTs and MT-associated structures made in the absence of two other TGB proteins remained uncertain the significance of MT localization for the TGBp1 intracellular movement. Previously, we analyzed TGBp2/TGBp3-dependent intra- and intercellular transport of PMTV GFP-TGBp1 using the experimental system based on particle bombardment with two expression vectors [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.]. This system had two major disadvantages. First, the relative amounts of the expression plasmids delivered in each particular cell can vary that may result in variations of the TGBp1 to TGBp2/TGBp3 ratio in different cells. Second, both the TGBp1 gene and the TGBp2/TGBp3 bicistronic gene region were cloned in expression cassettes under the control of the 35S promoter that results, taking into account that translation of a BSMV bicistronic template gives a TGBp2:TGBp3 ratio of 10:1 [20Zhou H, Jackson AO. Expression of the barley stripe mosaic virus RNA beta ‘triple gene block’ Virology 1996; 216: 367-79.], in an approximate TGBp1:TGBp2:TGBp3 molar ratio of 10:10:1 in co-bombarded cells instead of 100:10:1 ratio expected for natural virus infection on the basis of indirect data such as mRNA levels and in vitro translation efficiencies [9Jackson AO, Lim HS, Bragg J, Ganesan U, Lee MY. Hordeivirus replication, movement, and pathogenesis Annu Rev Phytopathol 2009; 47: 385-422.]. Therefore, both TGBp2 and TGBp3 were present in co-bombardment experiments in approximately ten-fold excess relative to TGBp1 amounts.
To improve the described system for further PMTV TGB studies we constructed a binary vector harboring two expressing cassettes, one of which is driven by the 35S promoter and contains GFP-fused PMTV TGBp1 gene, while another contains the TGBp2/TGBp3-encoding region cloned under the control of the promoter and transcription terminator of the A. tumefaciens nopaline synthase (NOS). The NOS promoter is known to be considerably weaker compared to the 35S promoter [30Sanders PR, Winter JA, Barnason AR, Rogers SG, Fraley RT. Comparison of cauliflower mosaic virus 35S and nopaline synthase promoters in transgenic plants Nucleic Acids Res 1987; 15: 1543-58.]. The two-cassette construct was termed pLH-35S:GFP-TGBp1-NOS:TGBp2/ TGBp3 (Fig. 2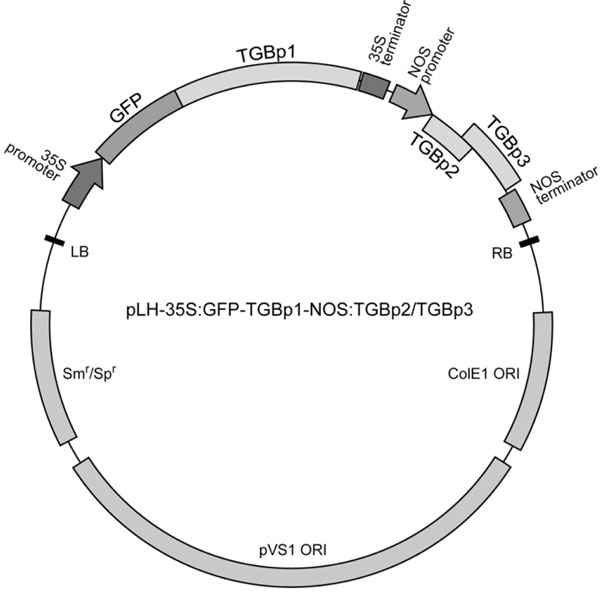 ).
).
Epidermal cells of N. benthamiana leaves agroinfiltrated with the construct 35S:GFP-TGBp1-NOS:TGBp2/TGBp3 were imaged using confocal laser scanning microscopy at different time points. At one day post infiltration (dpi) no visible GFP fluorescence was observed in infiltrated leaf areas (data not shown). At 2 dpi the GFP fluorescence was detected in the cytoplasm and within the nucleus as previously observed [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.]; the cytoplasmic fluorescence was mainly associated with MTs similar to those found in bombardment experiments (Fig. 3A ). At 3 dpi GFP-TGBp1 was again found in the nucleus and association with MT. Additionally, GFP-TGBp1 was detected in numerous granular bodies of 0.3 - 0.8 µm in size which were located both along the MTs and close to the cell wall (Fig. 3B
). At 3 dpi GFP-TGBp1 was again found in the nucleus and association with MT. Additionally, GFP-TGBp1 was detected in numerous granular bodies of 0.3 - 0.8 µm in size which were located both along the MTs and close to the cell wall (Fig. 3B , C
, C ). Using time-lapse microscopy we failed to detect movement of GFP-TGBp1-containing MT-associated granular structures in time periods of up to 1 min (data not shown). At 4 dpi, GFP-TGBp1 was detected mostly in cell wall-associated bodies and also in residual MTs, the nucleoplasm and large amorphous inclusions comparable in size to the nucleus (Fig. 3D
). Using time-lapse microscopy we failed to detect movement of GFP-TGBp1-containing MT-associated granular structures in time periods of up to 1 min (data not shown). At 4 dpi, GFP-TGBp1 was detected mostly in cell wall-associated bodies and also in residual MTs, the nucleoplasm and large amorphous inclusions comparable in size to the nucleus (Fig. 3D ). Typically, one such inclusion was found in each transformed cell close to the nucleus (Fig. 3D
). Typically, one such inclusion was found in each transformed cell close to the nucleus (Fig. 3D ). These structures resembled aggresomes which are formed in MT-dependent manner in response to excessive amounts of misfolded proteins in mammalian cells [31Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins J Cell Biol 1998; 143: 1883-98.] and described recently also for plants [32Vogel F, Hofius D, Sonnewald U. Intracellular trafficking of Potato leafroll virus movement protein in transgenic Arabidopsis Traffic 2007; 8: 1205-4.]. It should be noted that aggresome-like structures were observed in some cells also at 3 dpi (data not shown). Therefore, similarly to previous bombardment experiments with two expression vectors [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.], agrobacterium-mediated expression of PMTV TGBp1, TGBp2 and TGBp3 from a single binary vector resulted in targeting of GFP-TGBp1 to cell wall-associated structures. However, in the case of agroinfiltration, localization of GFP-TGBp1 to large membranous ‘intermediate’ bodies, or PMBs, described earlier [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.], was not observed. These structures therefore could have arisen due to TGBp2 and/or TGBp3 overexpression and represented artificial perturbations of cellular membranes [8Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. Varied movement strategies employed by triple gene block-encoding viruses Mol Plant-Microbe Interact 2010; 23: 1231-47.]. On the other hand, observations of infiltrated leaves at different time points revealed temporal localization of GFP-TGBp1 to different subcellular structures. Importantly, in these experiments association of GFP-TGBp1 with MT-like structures always preceded its detection in cell wall-associated sites.
). These structures resembled aggresomes which are formed in MT-dependent manner in response to excessive amounts of misfolded proteins in mammalian cells [31Johnston JA, Ward CL, Kopito RR. Aggresomes: a cellular response to misfolded proteins J Cell Biol 1998; 143: 1883-98.] and described recently also for plants [32Vogel F, Hofius D, Sonnewald U. Intracellular trafficking of Potato leafroll virus movement protein in transgenic Arabidopsis Traffic 2007; 8: 1205-4.]. It should be noted that aggresome-like structures were observed in some cells also at 3 dpi (data not shown). Therefore, similarly to previous bombardment experiments with two expression vectors [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.], agrobacterium-mediated expression of PMTV TGBp1, TGBp2 and TGBp3 from a single binary vector resulted in targeting of GFP-TGBp1 to cell wall-associated structures. However, in the case of agroinfiltration, localization of GFP-TGBp1 to large membranous ‘intermediate’ bodies, or PMBs, described earlier [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.], was not observed. These structures therefore could have arisen due to TGBp2 and/or TGBp3 overexpression and represented artificial perturbations of cellular membranes [8Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. Varied movement strategies employed by triple gene block-encoding viruses Mol Plant-Microbe Interact 2010; 23: 1231-47.]. On the other hand, observations of infiltrated leaves at different time points revealed temporal localization of GFP-TGBp1 to different subcellular structures. Importantly, in these experiments association of GFP-TGBp1 with MT-like structures always preceded its detection in cell wall-associated sites.
Co-Localization of GFP-TGBp1 with Plasmodesmata
To determine whether the GFP-TGBp1-containing cell wall-associated structures are related to plasmodesmata, plasmodesmata-associated callose was visualized by staining with sirofluor, a specific fluorescent dye [27Stone BA, Evans NA, Bonig I, Clarke AE. The application of sirofluor, a chemically defined fluorochrome from aniline blue for the histochemical detection of callose Protoplasma 1984; 122: 191-5.]. Leaves infiltrated with 35S:GFP-TGBp1-NOS:TGBp2/TGBp3 construct were stained at 3 dpi and analyzed by fluorescent microscopy. Two-color imaging revealed stained callose depositions along cell walls, and some of them coincided with GFP-TGBp1-containing structures (Fig. 4A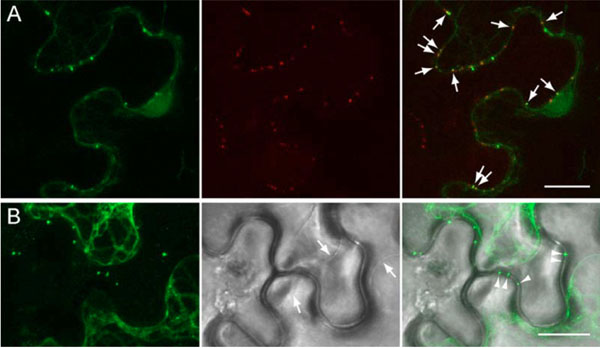 ). These data confirmed directed targeting of GFP-fused PMTV TGBp1 to plasmodesmata in the presence of TGBp2 and TGBp3.
). These data confirmed directed targeting of GFP-fused PMTV TGBp1 to plasmodesmata in the presence of TGBp2 and TGBp3.
Additionally, we analyzed whether the TGBp1-contained structures are peripherally associated with plasmodesmata or localize within cell wall-crossing microchannels and/or plasmodesmata central cavity. To this end, the portions of N. benthamiana leaves infiltrated with 35S:GFP-TGBp1-NOS:TGBp2/TGBp3 construct were subjected to plasmolysis by mannitol treatment, and epidermal cells were studied microscopically. After the treatment, the plasma membranes were retracted from cell walls, and gaps between them were visible in the bright field (Fig. 4B , arrows). Fluorescent microscopy revealed that after plasmolysis many GFP-TGBp1-containing bodies retained in cell walls (Fig. 4B
, arrows). Fluorescent microscopy revealed that after plasmolysis many GFP-TGBp1-containing bodies retained in cell walls (Fig. 4B , arrowheads). Thus, TGBp1-containing structures reside within cell walls, most probably - within plasmodesmata central cavity.
, arrowheads). Thus, TGBp1-containing structures reside within cell walls, most probably - within plasmodesmata central cavity.
Colchicine Treatment Abolishes TGBp1 Plasmodesmata Targeting
Upon leaf agroinfiltration with 35S:GFP-TGBp1-NOS:TGBp2/TGBp3 construct, GFP-TGBp1 association with MTs preceded to its localization to plasmodesmata. Therefore we verified whether interaction with MTs is required for TGBp1 intracellular transport to plasmodesmata. To this end we analyzed the transport of GFP-TGBp1 to plasmodesmata in cells where MTs were disrupted by colchicine. In this series of experiments, colchicine was added to an infiltration buffer prior to agroinfiltration of N. benthamiana leaves with 35S:GFP-TGBp1-NOS:TGBp2/TGBp3 construct. In parallel, control leaves were infiltrated with 35S:YFP-MAP4 and treated with colchicine. At 3 dpi, microscopy of YFP-MAP4-agroinfiltrated leaves treated with colchicine revealed diffused fluorescence in the cytoplasm and in the nucleus (Fig. 5B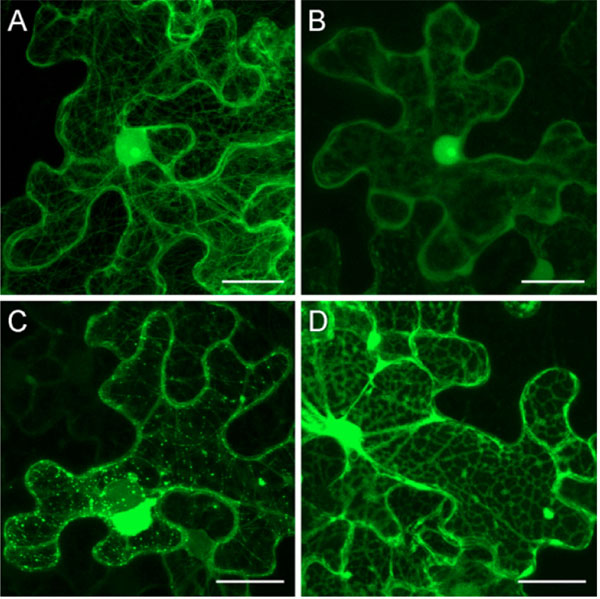 ). Additionally, as in non-treated leaves, YFP-MAP4 was detected in the nucleolus (Fig. 5A
). Additionally, as in non-treated leaves, YFP-MAP4 was detected in the nucleolus (Fig. 5A , B
, B ). In leaves agroinfiltrated with 35S:GFP-TGBp1-NOS:TGBp2/TGBp3 construct MTs, MT-associated bodies, plasmodesmata and aggresome-like structures visible in non-treated leaves at 3 dpi (Fig. 5C
). In leaves agroinfiltrated with 35S:GFP-TGBp1-NOS:TGBp2/TGBp3 construct MTs, MT-associated bodies, plasmodesmata and aggresome-like structures visible in non-treated leaves at 3 dpi (Fig. 5C ) were not observed in colchicine-treated leaves at 3 dpi (Fig. 5D
) were not observed in colchicine-treated leaves at 3 dpi (Fig. 5D ) and upon further observations at 4 dpi (data not shown). In these samples GFP-TGBp1 was found in the cytoplasm and in the nucleus. Surprisingly, GFP-TGBp1 was localized in the cytoplasm not diffusely but in association with an irregular network located in cell periphery and closely resembling the cortical endoplasmic reticulum (ER) (Fig. 5D
) and upon further observations at 4 dpi (data not shown). In these samples GFP-TGBp1 was found in the cytoplasm and in the nucleus. Surprisingly, GFP-TGBp1 was localized in the cytoplasm not diffusely but in association with an irregular network located in cell periphery and closely resembling the cortical endoplasmic reticulum (ER) (Fig. 5D ). Further experiments are being carried out to analyze the TGBp1 potential to interact with ER membranes in different experimental conditions. Whatever the nature of the observed ER-like structures, our data show that disassembly of cell MTs by colchicine prevented GFP-TGBp1 targeting to plasmodesmata and the MT-dependent aggresome formation.
). Further experiments are being carried out to analyze the TGBp1 potential to interact with ER membranes in different experimental conditions. Whatever the nature of the observed ER-like structures, our data show that disassembly of cell MTs by colchicine prevented GFP-TGBp1 targeting to plasmodesmata and the MT-dependent aggresome formation.
Deletion Analysis Reveals Correlation Between Microtubule Association and Plasmodesmata Targeting
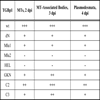
To identify protein regions involved in TGBp1 association with MTs, a series of TGBp1 mutants was constructed. They included four deletion mutants containing deletions in the TGBp1 N-terminal extension region, dN with the deletion of residues 1-34 including a stretch of positively charged residues, Mlu1 with deletion of residues 37-85 described previously [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.], Mlu2 with deletion of residues 37-184, and HEL with deletion of residues 1-187 (Fig. 6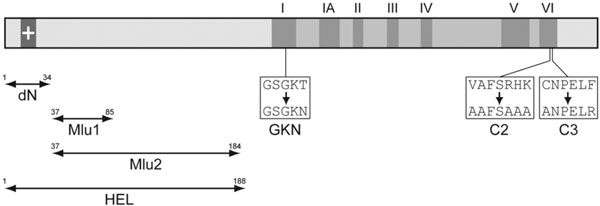 ). Additionally, three point mutants were constructed, GKN with T to N substitution in the conserved motif I of the TGBp1 NTPase/helicase domain, and mutants C2 and C3 with substitutions in the motif VI (Fig. 6
). Additionally, three point mutants were constructed, GKN with T to N substitution in the conserved motif I of the TGBp1 NTPase/helicase domain, and mutants C2 and C3 with substitutions in the motif VI (Fig. 6 ). T to N mutations in NTPase motif 1 (Walker A nucleotide-binding site) are well documented to result in much higher affinity of NTPases for ADP/GDP than for ATP/GTP thus disabling energy conversion even in the presence of ATP and blocking therefore the protein enzymatic/regulatory functions [33Feig LA. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases Nat Cell Biol 1999; 1: E25-7., 34Nagy M, Wu HC, Liu Z, Kedzierska-Mieszkowska S, Zolkiewski M. Walker-A threonine couples nucleotide occupancy with the chaperone activity of the AAA+ ATPase ClpB Protein Sci 2009; 18: 287-93.]. Mutation introduced in C2 and C3 fall into a region potentially involved in interactions with TGBp2 or TGBp3 which are necessary for the TGBp2/TGBp3-directed targeting of TGBp1 to plasmodesmata [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.]. The mutant TGBp1 genes were cloned into pLH-35S:GFP-TGBp1-NOS:TGBp2/TGBp3 to replace the wild type sequence, the resulting vectors were transformed into A. tumifaciens, and the obtained agrobacterial cultures were used for infiltration of N. benthamiana leaves. Association of mutants with sub-cellular structures was analyzed by microscopy at 2, 3 and 4 dpi and compared to that of the wild-type protein. It was found that the mutations dN, Mlu1, GKN, C2 and C3 had, to different extent, a suppressive effect on the GFP-TGBp1 ability to interact with MTs, to localize to MT-associated bodies and to be targeted to plasmodesmata; however this group of mutants retained the ability of wild-type protein to interact with these cell sub-structures (Table 1). In contrast, the mutants Mlu2 and HEL completely lost the ability for association with MTs and plasmodesmata targeting (Table 1; Fig. 7
). T to N mutations in NTPase motif 1 (Walker A nucleotide-binding site) are well documented to result in much higher affinity of NTPases for ADP/GDP than for ATP/GTP thus disabling energy conversion even in the presence of ATP and blocking therefore the protein enzymatic/regulatory functions [33Feig LA. Tools of the trade: use of dominant-inhibitory mutants of Ras-family GTPases Nat Cell Biol 1999; 1: E25-7., 34Nagy M, Wu HC, Liu Z, Kedzierska-Mieszkowska S, Zolkiewski M. Walker-A threonine couples nucleotide occupancy with the chaperone activity of the AAA+ ATPase ClpB Protein Sci 2009; 18: 287-93.]. Mutation introduced in C2 and C3 fall into a region potentially involved in interactions with TGBp2 or TGBp3 which are necessary for the TGBp2/TGBp3-directed targeting of TGBp1 to plasmodesmata [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.]. The mutant TGBp1 genes were cloned into pLH-35S:GFP-TGBp1-NOS:TGBp2/TGBp3 to replace the wild type sequence, the resulting vectors were transformed into A. tumifaciens, and the obtained agrobacterial cultures were used for infiltration of N. benthamiana leaves. Association of mutants with sub-cellular structures was analyzed by microscopy at 2, 3 and 4 dpi and compared to that of the wild-type protein. It was found that the mutations dN, Mlu1, GKN, C2 and C3 had, to different extent, a suppressive effect on the GFP-TGBp1 ability to interact with MTs, to localize to MT-associated bodies and to be targeted to plasmodesmata; however this group of mutants retained the ability of wild-type protein to interact with these cell sub-structures (Table 1). In contrast, the mutants Mlu2 and HEL completely lost the ability for association with MTs and plasmodesmata targeting (Table 1; Fig. 7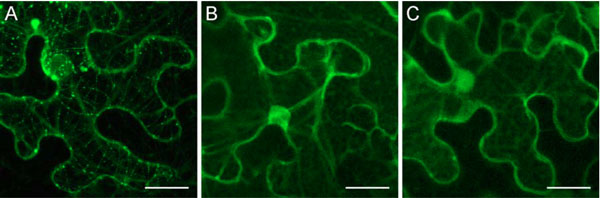 ). These data show the correlation between these two interactions characteristic of PMTV TGBp1 and support the colchicine inhibition data pointing to the dependence of TGBp1 plasmodesmata targeting on the integrity of cell MTs. Additionally, one can conclude that the TGBp1 region comprising residues 86 - 184 is involved in the protein association with MTs. This region includes a C-terminal portion of the TGBp1 N-terminal natively unfolded domain and most of the internal structured domain [10Makarov VV, Rybakova EN, Efimov AE, et al. Domain organization of the N-terminal portion of hordeivirus movement protein TGBp1 J Gen Virol 2009; 90: 3022-2.]. Amino acid sequence alignment of TGBp1s encoded by four pomoviruses revealed that this region exhibits considerable sequence conservation in its C-terminal portion corresponding to the protein internal domain, whereas the N-terminal domain appeared to be weakly conserved even among these closely related viruses (Fig. 8
). These data show the correlation between these two interactions characteristic of PMTV TGBp1 and support the colchicine inhibition data pointing to the dependence of TGBp1 plasmodesmata targeting on the integrity of cell MTs. Additionally, one can conclude that the TGBp1 region comprising residues 86 - 184 is involved in the protein association with MTs. This region includes a C-terminal portion of the TGBp1 N-terminal natively unfolded domain and most of the internal structured domain [10Makarov VV, Rybakova EN, Efimov AE, et al. Domain organization of the N-terminal portion of hordeivirus movement protein TGBp1 J Gen Virol 2009; 90: 3022-2.]. Amino acid sequence alignment of TGBp1s encoded by four pomoviruses revealed that this region exhibits considerable sequence conservation in its C-terminal portion corresponding to the protein internal domain, whereas the N-terminal domain appeared to be weakly conserved even among these closely related viruses (Fig. 8 ). This observation suggests that the PMTV TGBp1 internal domain can be involved in specific interactions with MTs.
). This observation suggests that the PMTV TGBp1 internal domain can be involved in specific interactions with MTs.
DISCUSSION
In this paper we report co-localization of GFP-fused PMTV TGBp1 with MTs. Thus, TGBp1 can be added to a list of plant virus MPs capable of interaction, direct or indirect, with MTs. The most extensively studied MP encoded by TMV is among these proteins. During infection, TMV MP interacts with endoplasmic reticulum (ER) membranes and MTs, as well as localizes to plasmodesmata and induces an increase of their size exclusion limit [3Taliansky M, Torrance L, Kalinina NO. Role of plant virus movement proteins Methods Mol Biol 2008; 451: 33-54., 4Epel BL. Plant viruses spread by diffusion on ER-associated movement-protein-rafts through plasmodesmata gated by viral induced host beta-1, 3-glucanases Semin Cell Dev Biol 2009; 20: 1074-81., 35Heinlein M, Epel BL. Macromolecular transport and signaling through plasmodesmata Int Rev Cytol 2004; 235: 93-164.]. TMV MP also has the capacity to bind single-stranded nucleic acids in vitro, and the movement of viral RNA genome likely occurs in the form of vRNP [3Taliansky M, Torrance L, Kalinina NO. Role of plant virus movement proteins Methods Mol Biol 2008; 451: 33-54.]. Thus TMV MP shares with PMTV TGBp1 at least two activities involved in formation of vRNPs and interaction with cortical MTs [8Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. Varied movement strategies employed by triple gene block-encoding viruses Mol Plant-Microbe Interact 2010; 23: 1231-47., 35Heinlein M, Epel BL. Macromolecular transport and signaling through plasmodesmata Int Rev Cytol 2004; 235: 93-164.]. TMV MP sequences required for interaction with MTs are identified by mutation analyses [36Kahn TW, Lapidot M, Heinlein M, et al. Domains of the TMV movement protein involved in subcellular localization Plant J 1998; 15: 15-25., 37Boyko V, van der Laak J, Ferralli J, Suslova E, Kwon MO, Heinlein M. Cellular targets of functional and dysfunctional mutants of tobacco mosaic virus movement protein fused to green fluorescent protein J Virol 2000; 74: 11339-46.]. Moreover, a region within TMV MP is revealed that shares sequence similarity to tubulins, and single amino acid substitutions within this so-called ‘tubulin mimicry domain’ are sufficient to confer temperature-sensitive loss of MT association pointing to the possibility that TMV MP is capable of making binding contacts equivalent to tubulin-tubulin interaction [38Boyko V, Ferralli J, Ashby J, Schellenbaum P, Heinlein M. Function of microtubules in intercellular transport of plant virus RNA Nat Cell Biol 2000; 2: 826-32.]. These data, together with biochemical evidences obtained in vitro [39McLean B, Zupan J, Zambryski P. Tobacco mosaic movement protein associates with the cytoskeleton in tobacco cells Plant Cell 1995; 7: 2101-14.-41Sambade A, Brandner K, Hofmann C, Seemanpillai M, Mutterer J, Heinlein M. Transport of TMV movement protein particles associated with the targeting of RNA to plasmodesmata Traffic 2008; 9: 2073-88.], indicate that TMV MP can interact with tubulin and MTs directly. Although we were unable to find a sequence similar to the ‘tubulin mimicry domain’ in PMTV TGBp1 (data not shown), deletion analysis suggest that the TGBp1 region encompassing residues 86 - 184 is involved in MT association. Assuming that the predicted subdomain organization of PMTV TGBp1 is very similar to that of hordeivirus TGBp1 [10Makarov VV, Rybakova EN, Efimov AE, et al. Domain organization of the N-terminal portion of hordeivirus movement protein TGBp1 J Gen Virol 2009; 90: 3022-2.], one can conclude that among three domains of the hordei-like TGBp1s the structured ID domain is responsible for the protein association with MTs. Taking into account our preliminary data indicating that PMTV TGBp1 can interact with tobacco tubulin A1 in vitro, we presume the direct physical TGBp1 interaction with MTs.
What is the possible significance of the targeting of PMTV TGBp1 to the MTs? Recent findings illuminated the role of MTs in the intracellular transport of TMV vRNPs. Using a TMV derivative encoding a temperature-sensitive TMV MP, small mobile MT-proximal particles representing MP-RNA complexes were revealed in cells at the leading front of infection [41Sambade A, Brandner K, Hofmann C, Seemanpillai M, Mutterer J, Heinlein M. Transport of TMV movement protein particles associated with the targeting of RNA to plasmodesmata Traffic 2008; 9: 2073-88., 42Boyko V, Hu Q, Seemanpillai M, Ashby J, Heinlein M. Validation of microtubule-associated tobacco mosaic virus RNA movement and involvement of microtubule-aligned particle trafficking Plant J 2007; 51: 589-603.]. Importantly, the TMV MP-containing particles formed upon MP transient expression in the absence of viral infection are functionally related to the particles observed in virus-infected cells; these particles contain RNA and are thought to be assembled on the MT network; their anchorage to MT sites and release can contribute to the intracellular transport of these specific RNPs to plasmodesmata [41Sambade A, Brandner K, Hofmann C, Seemanpillai M, Mutterer J, Heinlein M. Transport of TMV movement protein particles associated with the targeting of RNA to plasmodesmata Traffic 2008; 9: 2073-88.]. Similarly to TMV MP, PMTV TGBp1 was also found in granular bodies associated with MTs. However, in contrast to the TMV MP, we were unable to detect movement of TGBp1-containing bodies. Nevertheless, association of TGBp1 with MTs and its localization in MT-associated granular bodies can be necessary for the protein targeting to plasmodesmata in the presence of TGBp2 and TGBp3, since treatment with the microtubule-disrupting drug, colchicine, inhibits localization of TGBp1 to plasmodesmata. One possible function of TGBp1 association with MTs is that the MT network may provide a scaffold for assembly of TGBp1-containing granular bodies as an initial step in TGBp1 targeting to plasmodesmata by TGBp2 and TGBp3. It should be noted that the Cauliflower mosaic virus proteins P2, P3 and P6 also form bodies that localize to MTs and, moreover, require intact MTs for their efficient formation [43Harries PA, Palanichelvam K, Yu W, Schoelz JE, Nelson RS. The cauliflower mosaic virus protein P6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules Plant Physiol 2009; 149: 1005-6., 44Martinière A, Gargani D, Uzest M, Lautredou N, Blanc S, Drucker M. A role for plant microtubules in the formation of transmission-specific inclusion bodies of Cauliflower mosaic virus Plant J 2009; 58: 135-46.], lending support to the possibility that a similar mechanism may be utilized for the formation of MT-associated granular structures containing PMTV TGBp1.
Additionally, localization to MTs could facilitate TGBp1 degradation. The degradation of Potato leafroll virus MP, for example, was found to require intact MTs, and a similar model has been proposed for the MT-associated degradation of TMV MP [32Vogel F, Hofius D, Sonnewald U. Intracellular trafficking of Potato leafroll virus movement protein in transgenic Arabidopsis Traffic 2007; 8: 1205-4., 45Curin M, Ojangu EL, Trutnyeva K, Ilau B, Truve E, Waigmann E. MPB2C, a microtubule-associated plant factor, is required for microtubular accumulation of tobacco mosaic virus movement protein in plants Plant Physiol 2007; 143: 801-11.]. It was suggested that individual MP molecules or their small aggregates could traffic along MTs to stationary sites of the formation of aggresome-like structures [32Vogel F, Hofius D, Sonnewald U. Intracellular trafficking of Potato leafroll virus movement protein in transgenic Arabidopsis Traffic 2007; 8: 1205-4.]. Indeed, a similar model has been proposed for the formation of aggresomes, perinuclear structures formed in animal cells in response to protein overproduction and misfolding [46Garcia-Mata R, Gao YS, Sztul E. Hassles with taking out the garbage: aggravating aggresomes Traffic 2002; 3: 388-96.]. Evidently, transient expression of PMTV TGBp1 alone or together with TGBp2 and TGBp3 induced formation of typical single perinuclear aggresome-like structure in transformed cell. Moreover, colchicine blocked both TGBp1 localization to plasmodesmata and formation of perinuclear aggresomes.
Current view of TMV MP intracellular transport is based on the fact that ER membrane proteins can laterally diffuse within the membrane [47Runions J, Brach T, Kühner S, Hawes C. Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane J Exp Bot 2006; 57: 43-50.] and move from cell to cell [4Epel BL. Plant viruses spread by diffusion on ER-associated movement-protein-rafts through plasmodesmata gated by viral induced host beta-1, 3-glucanases Semin Cell Dev Biol 2009; 20: 1074-81., 46Garcia-Mata R, Gao YS, Sztul E. Hassles with taking out the garbage: aggravating aggresomes Traffic 2002; 3: 388-96.]. Since the ER is tightly associated with the actin network [49Boevink P, Oparka K, Sant Cruz S, Martin B, Betteridge A, Hawes C. Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network Plant J 1998; 15: 441-7.], microfilaments could play an important role in supporting the ER-mediated targeting of MP and/or vRNA to plasmodesmata. Assuming that in plant cells MTs and actin cables may co-align at cell periphery [50Barton DA, Overall RL. Cryofixation rapidly preserves cytoskeletal arrays of leaf epidermal cells revealing microtubule co-alignments between neighbouring cells and adjacent actin and microtubule bundles in the cortex J Microsc 2010; 237: 79-88.] it can be proposed that the MP or viral transport RNP, initially anchored to MT-proximal sites, may be able to detach and undergo movements by an ER-driven mechanism [41Sambade A, Brandner K, Hofmann C, Seemanpillai M, Mutterer J, Heinlein M. Transport of TMV movement protein particles associated with the targeting of RNA to plasmodesmata Traffic 2008; 9: 2073-88.]. Our data indirectly support applicability of a similar model to the PMTV transport system. After the colchicine treatment GFP-TGBp1 was localized in the cytoplasm not diffusely but in association with an irregular network closely resembling the cortical ER, suggesting its potential ER binding ability. Although the nature of these ER-like structures remains to be established, one can speculate that TGBp1 association with the ER could be due to integral membrane proteins TGBp2 and/or TGBp3 associated with the ER or ER-derived membrane compartments [7Morozov SY, Solovyev AG. Triple gene block: modular design of a multifunctional machine for plant virus movement J Gen Virol 2003; 84: 1351-66.].
CONCLUSION
To conclude, we suggest the model of PMTV TGBp1 functioning emphasizing the important role of MTs. Early in infection TGBp1 can laterally associate with MTs to form MT-associated granular bodies. After some amounts of TGBp2 integrated into ER membranes are accumulated in infected cell, TGBp1-containing granular bodies, due to the suggested TGBp1 ability to interact with TGBp2 [16Zamyatnin AA Jr, Solovyev AG, Savenkov EI, et al. Transient coexpression of individual genes encoded by the triple gene block of potato mop-top virus reveals requirements for TGBp1 trafficking Mol Plant-Microbe Interact 2004; 17: 921-30.], are directed to the sites of TGBp2 localization at close contacts between MTs and ER. It can be speculated that the TGBp1-containing granular bodies include also RNA because of RNA binding activities of TGBp1 and TGBp2 [8Verchot-Lubicz J, Torrance L, Solovyev AG, Morozov SY, Jackson AO, Gilmer D. Varied movement strategies employed by triple gene block-encoding viruses Mol Plant-Microbe Interact 2010; 23: 1231-47.]. Increase of TGBp3 amount results in a rapid ER-driven movement of TGBp1/TGBp2/RNA complexes to plasmodesmata and through plasmodesmata to neighboring cells. Further increase of concentration of MT-bound TGBp1 results in its targeting to aggresomes for degradation that can be important at the later steps of infection cycle when virions are forming, and excessive amounts of RNA-binding protein TGBp1 can compete with the coat protein for viral genomic RNAs. Thus, we propose that TGBp1 interaction with MTs may be important for both the formation of vRNP bodies early in infection and degradation of the excessive TGBp1 at late infection stages.
ACKNOWLEDGEMENTS
We are grateful to DAAD (German Academic Exchange Service) for the short-term fellowship awarded to Elena Shemyakina to support her research work at Joachim Schiemann laboratory at Julius Kühn Institute.