- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Role of Filopodia in HSV-1 Entry into Zebrafish 3-O-Sulfotransferase-3-Expressing Cells
Samiksha Choudhary 1, Lorrie Burnham 2, Jeffrey M Thompson 3, Deepak Shukla 4, Vaibhav Tiwari*, 5
Abstract
Background:
Heparan sulfate proteoglycans (HSPGs) modified by zebrafish (ZF) encoded glucosaminyl 3-O sulfotransferase-3 (3-OST-3) generate a receptor for herpes simplex virus type-1 (HSV-1) entry and spread. In order to elucidate the mechanism by which HSV-1 enters into ZF-3-OST-3 cells, we investigated the mode of viral entry.
Results:
Under high resolution scanning electron microscopy (SEM), actin cytoskeleton changes were observed by a dramatic increase in the number of filopodia formed during early interactions of HSV-1 with the target cells. While the increase in number was common among all the infected cells, the highest numbers of filopodia was observed in cells expressing the 3-OST-3 modified form of heparan sulfate (HS) encoded either by human or ZF. The levels of viral infection and filopodia induction were reduced with the actin polymerization inhibitors, Cytochalasin-D and Lantriculin B, suggesting an important role for actin reorganization during ZF-3-OST-3 mediated HSV-1 entry. Supporting an interesting possibility of filopodia usage during HSV-1 spread, pre-treatment of cytochalasin D in ZF-3-OST-3 cells drastically reduced virus glycoprotein induced cell fusion.
Conclusions:
Taken together, our results provide new evidence on the involvement of filopodia during HSV-1 infection of ZF-3-OST-3 cells and confirm a role for modified heparan sulfate in cytoskeleton rearrangement during HSV-1 entry.
Article Information
Identifiers and Pagination:
Year: 2013Volume: 7
First Page: 41
Last Page: 48
Publisher Id: TOVJ-7-41
DOI: 10.2174/1874357901307010041
Article History:
Received Date: 20/11/2012Revision Received Date: 11/1/2013
Acceptance Date: 23/1/2013
Electronic publication date: 5 /4/2013
Collection year: 2013
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Department of Microbiology & Immunology, Midwestern University, Downers Grove IL 60515, USA; Fax: + 1 630-515-6358; E-mail: vtiwar@midwestern.edu
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 20-11-2012 |
Original Manuscript | Role of Filopodia in HSV-1 Entry into Zebrafish 3-O-Sulfotransferase-3-Expressing Cells | |
BACKGROUND
Heparan sulfate (HS) is a glycosaminoglycan (GAG) of repeating disaccharide units that consist of N-acetylglucos-amine and glucuronic/iduoronic acid [1Esko JD, Lindahl U. Molecular diversity of heparan sulfate J Clin Invest 2001; 108: 169-73., 2Lindahl U, Kusche-Gullberg M, Kjellen L. Regulated diversity of heparan sulfate J Biol Chem 1998; 273: 24979-82.]. They are expressed on the cell surface and in the extracellular matrix as a hybrid molecule of heparan sulfate proteoglycans (HSPGs) attached to a protein core which mediate crucial functions ranging from cell adhesion, cell signaling, blood coagulation, to wound healing and growth-factor mediated proliferation [3Gorsi B, Stringer SE. Tinkering with heparan sulfate sulfation to steer development Trends Cell Biol 2007; 17: 173-7.-5Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine tune mammalian physiology Nature 2007; 446: 1030-7-0.]. The long chains of HS containing alternating disaccharides are modified in a complex series of steps during its synthesis. In each of the modification steps only part of the substrate is modified resulting in high sequence diversity, which is thought to give HSPGs their functional specificity and versatility. Thus, each 3-OST can potentially generate unique protein-binding sites within HS chain [1Esko JD, Lindahl U. Molecular diversity of heparan sulfate J Clin Invest 2001; 108: 169-73., 6Shukla D, Spear PG. Herpesviruses and heparan sulfate an intimate relationship in aid of viral entry J Clin Invest 2001; 108: 503-10.].
The current model of herpes simplex virus type-1 (HSV-1) entry suggests multiple roles for both HS and modified HS during entry and spread [7Spear PG, Longnecker R. Herpesvirus entry an update J Virol 2003; 77: 10179-85.-9O'Donnell CD, Kovacs M, Akhtar J, Valyi-Nagy T, Shukla D. Expanding the role of 3-O sulfated heparan sulfate in herpes simplex virus type-1 entry Virology 2010; 397: 389-98.]. For instance, the unmodified form of HS mediates the initial viral binding or attachment to the host cells via HSV-1 glycoprotein B (gB) [10WuDunn D, Spear PG. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate J Virol 1989; 63: 52-8.] followed by the fusion between viral envelope glycoprotein D (gD) and the host cell membrane during viral spread [11Shukla D, Liu J, Blaiklock P, et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry Cell 1999; 99: 13-22.]. In the latter process of fusion it is the 3-O-sulfotransferase enzymatic modification of HS chains that are involved [11Shukla D, Liu J, Blaiklock P, et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry Cell 1999; 99: 13-22.-15Xu D, Tiwari V, Xia G, Clement C, Shukla D, Liu J. Characterization of heparan sulphate 3-O-sulphotransferase isoform 6 and its role in assisting the entry of herpes simplex virus type 1 Biochem J 2005; 385: 451-9.]. Emerging evidence further suggests that HS also assists HSV-1 surfing on filopodia to reach cell body for successful infection [16Oh MJ, Akhtar J, Desai P, Shukla D. A role for heparan sulfate in viral surfing Biochem Biophys Res Commun 2010; 391: 176-81.].
3-OST enzymes that modify HS play an essential role during zebrafish (ZF) embryo development [17Bink RJ, Habuchi H, Lele Z, et al. Heparan sulfate 6-O-sulfotransferase is essential for muscle development in zebrafish J Biol Chem 2003; 278: 31118-27.-20Liu IH, Zhang C, Kim MJ, Cole GJ. Retina development in zebrafish requires the heparan sulfate proteoglycan Agrin Dev Neurobiol 2008; 68()(7 ): 877-98.]. The expression patterns of multiple isoforms of 3-OST and their significance were reported in ZF [21Cadwallader AB, Yost HJ. Combinatorial expression patterns of heparan sulfate sulfotransferase in zebrafish I The 3-O-sulfotransferase family Dev Dynam 2006; 235: 3423-1.]. Recently, we reported that the ZF encoded 3-OST-3 isoform allows HSV-1 entry similar to the human 3-OST-3 isoform [22Hubbard S, Darmani NA, Thrush GR, et al. Zebrafish-encoded 3-O-sulfotransferase-3 isoform mediates herpes simplex virus type 1 entry and spread Zebrafish 2010; 7: 181-7.]. However, the mechanism by which HSV-1 entry in ZF encoded 3-OST-3 cells is facilitated remains to be defined. Therefore, in this study, we examined the mechanism of HSV-1 entry via ZF encoded 3-OST-3 isoform. The results presented below demonstrate that HSV-1 entry into ZF encoded 3-OST-3 receptor cells is facilitated by filopodia. The significance of the actin cytoskeleton in the ZF 3-OST-3 isoform is likely to open exciting new areas of investigations. This information will be important for developing a clear understanding of HSV-1 invasion of the neurons in the established ZF model of HSV-1 infection [23Burgos JS, Gomez JR, Alfaro JM, Sastre I, Valdivieso F. Zebrafish as a new model for herpes simplex virus type 1 infection Zebrafish 2008; 5: 323-3.]. Likewise, studying the role of 3-OST-3 modified HS in ZF infection model may also contribute to our understanding of inflammation and associated pathology [24Parish CR. The role of heparan sulphate in inflammation Nature Rev Immunol 2006; 6: 633-43.-26Aquino RS, Lee ES, Park PW. Diverse functions of glycosaminoglycans in infectious diseases Prog Mol Biol Transl Sci 2010; 93: 373-94.]. ZF has already been recognized for its value as a model for infectious diseases, inflammation, and immunity [27Sullivan C, Kim CH. Zebrafish as a model for infectious disease and immune function Fish Shellfish Immunol 2008; 25: 341-50.].
RESULTS
CHO-K1 Cells Expressing ZF 3-OST-3 Isoform Induce Membrane Protrusions During HSV-1 Entry
The previous studies have shown that primary cultures of corneal fibroblasts (CF) expressing the human encoded form of 3-OST-3 receptor leads to a change in the actin cytoskeleton during HSV-1 entry [30Clement C, Tiwari V, Scanlon PM, Vali-Nagy T, Yue BYJT, Shukla D. A novel for phagocytosis-like uptake in herpes simplex virus entry
J Cell Biol 2006; 174: 1009-21., 31Tiwari V, Clement C, Xu D, et al. Role for 3-O-sulfated heparan sulfate as a receptor for herpes simplex virus type-1 entry into primary human corneal fibroblasts
J Virol 2006; 80: 8970-0.]. Therefore, we examined whether cytoskeletal changes played any significant role in HSV-1 entry into CHO-K1 cells expressing ZF encoded 3-OST-3 isoform. To address this, a high resolution scanning electron microscopy (SEM) was performed. Wild-type HSV-1 (KOS) was added to the cells plated at a low population density for 30 min at 37oC. SEM images demonstrated that the infected cells expressing ZF encoded 3-OST-3 had numerous virions attached to the cell body while many others were also attached to filopodia-like projections present on the plasma membrane of ZF-3-OST-3 cell (Fig. 1A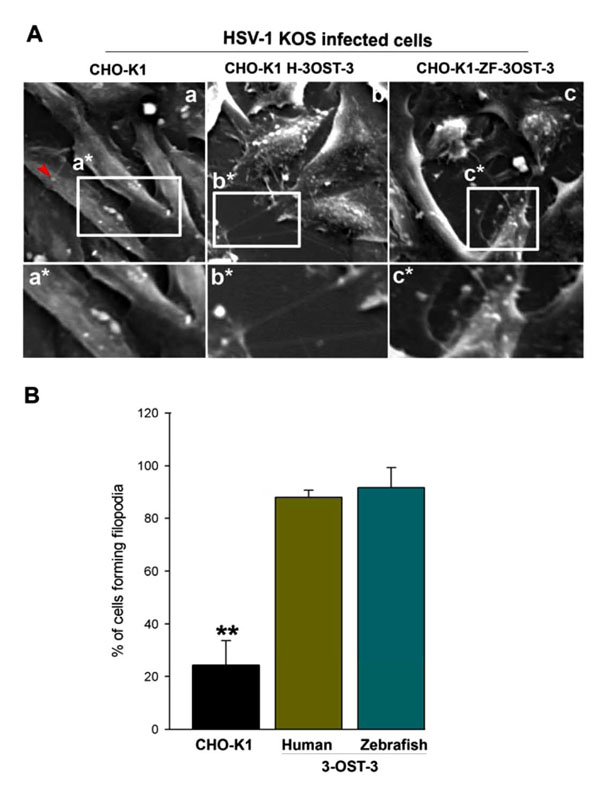 , panel c). Similar projections were also noticed in human 3-OST-3 isoform expressing CHO-K1 cells (Fig. 1A
, panel c). Similar projections were also noticed in human 3-OST-3 isoform expressing CHO-K1 cells (Fig. 1A , panel b). CHO-K1 cells without 3-OST-s expression showed little filopodia induction (Fig. 1A
, panel b). CHO-K1 cells without 3-OST-s expression showed little filopodia induction (Fig. 1A , panel a). The quantification on the number of filopodia formed in ZF-3-OST-3 cells was found to be comparable to the human 3-OST-3 expressing CHO-K1 cells (Fig. 1B
, panel a). The quantification on the number of filopodia formed in ZF-3-OST-3 cells was found to be comparable to the human 3-OST-3 expressing CHO-K1 cells (Fig. 1B ). The above data indicates a role of ZF 3-OST-3 in filopodia induction during HSV-1 entry.
). The above data indicates a role of ZF 3-OST-3 in filopodia induction during HSV-1 entry.
Enhanced Filopodia Induction is Seen in the Cells Expressing ZF Encoded 3-OST-3 Isoform
Because HSV-1 was able to induce filopodia both in wild-type CHO-K1 cells and CHO-K1 cells expressing ZF 3-OST-3, we next evaluated whether changes in actin cytoskeleton were dependent on HS. In order to generate direct and visual evidence of filopodia induction during HSV-1 entry we used SEM. In this experiment HSV-1 (KOS) was used to infect pCDNA3.1 expressing CHO-K1 cells and ZF-3-OST-3 expressing CHO-K1 cells and the above infections was compared for filopodia induction to CHO cells that are deficient in HS (CHO-745). As shown in Fig. (2A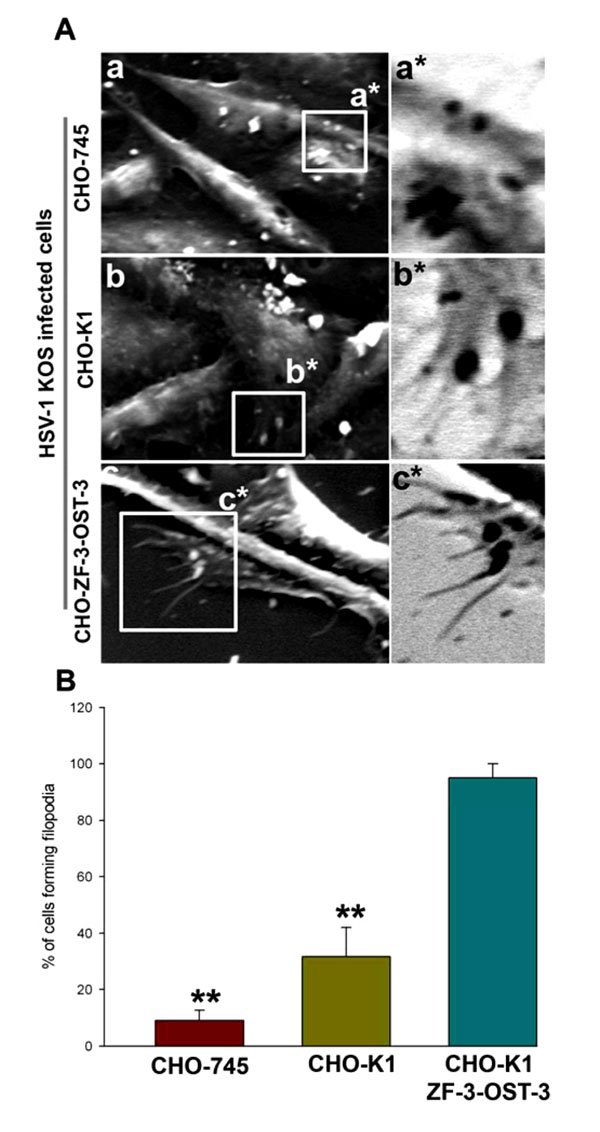 , panel a) HSV-1 infected CHO-745 (HS deficient) cells failed to produce significant amount of filopodia compared to the HS expressing wild-type CHO-K1 cells (Fig 2A
, panel a) HSV-1 infected CHO-745 (HS deficient) cells failed to produce significant amount of filopodia compared to the HS expressing wild-type CHO-K1 cells (Fig 2A , panel b). In parallel, pronounced filopodia were observed in CHO-K1 cells expressing ZF encoded 3-OST-3 isoform (Fig. 2A
, panel b). In parallel, pronounced filopodia were observed in CHO-K1 cells expressing ZF encoded 3-OST-3 isoform (Fig. 2A , panel c). Quantification of percentage of cells forming filopodia was highest in ZF-3-OST-3 cells (Fig. 2B
, panel c). Quantification of percentage of cells forming filopodia was highest in ZF-3-OST-3 cells (Fig. 2B ). These results indicate that filopodia induction during HSV-1 entry is HS dependent; Furthermore, the presence of 3-OST-3-modified receptor significantly enhances filopodia induction to facilitate HSV-1 entry into ZF expressing 3-OST-3 CHO-K1 cells.
). These results indicate that filopodia induction during HSV-1 entry is HS dependent; Furthermore, the presence of 3-OST-3-modified receptor significantly enhances filopodia induction to facilitate HSV-1 entry into ZF expressing 3-OST-3 CHO-K1 cells.
Role for Actin Cytoskeleton During HSV-1 Entry into Cells Expressing ZF 3-OST-3
To further demonstrate the significance of actin network in HSV-1 entry, both ZF and human 3-OST-3 receptor expressing CHO-K1 cells were pre-treated with inhibitors of actin polymerization such as cytochalasin D (Cyto D) and lantriculin B (Lat B) [32Gottlieb TA, Ivanov IE, Adesnik M, Sabatini DD. Actin microfilaments play a critical role in endocytosis at the apical but not the basolateral surface of polarized epithelial cells
J Cell Biol 1993; 120: 695-710.-34Schafer DA. Regulating actin dynamics at membranes a focus on dynamin
Traffic 2004; 5: 463-9.] or mock treated before infecting with Lac Z encoded β-galactosidase expressing HSV-1. It was postulated that pre-treatment would have negative effect on entry provided the actin-based protrusions (such as filopodia) played a role in the virus attachment as well as entry in to cells. As shown in Fig. (3A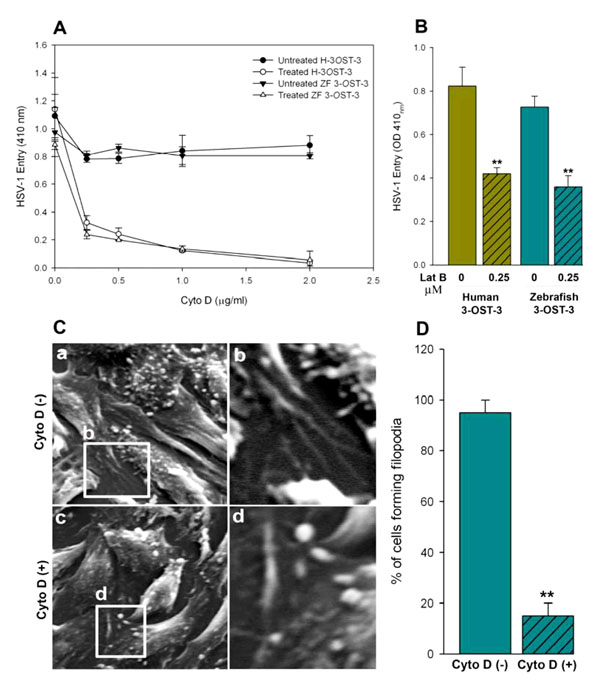 , B
, B ), both of the actin depolymerizers blocked HSV-1 entry in a dosage dependent manner. To generate further visual evidence, SEM was performed in HSV-1 infected ZF-3-OST-3 expressing CHO-K1 cells in presence and absence of Cyto D. Corresponding effects on the inhibition on the number of filopodia and HSV-1 entry was noticed when cells were pre-incubated with Cyto-D (Fig. 3C
), both of the actin depolymerizers blocked HSV-1 entry in a dosage dependent manner. To generate further visual evidence, SEM was performed in HSV-1 infected ZF-3-OST-3 expressing CHO-K1 cells in presence and absence of Cyto D. Corresponding effects on the inhibition on the number of filopodia and HSV-1 entry was noticed when cells were pre-incubated with Cyto-D (Fig. 3C , D
, D ). The quantitation of the above-mentioned data suggests that significant changes in the cytoskeleton occur during initial phase of HSV-1 infection in ZF encoded 3-OST-3 receptor. A similar increase in cellular projections was previously reported with other HSV-1 receptors including nectin-1 [35Tiwari V, Oh MJ, Kovacs M, Shukla SY, Valyi-Nagy T, Shukla D. Role for nectin-1 in herpes simplex virus 1 entry and spread in human retinal pigment epithelial cells
FEBS J 2008; 275: 5272-85.].
). The quantitation of the above-mentioned data suggests that significant changes in the cytoskeleton occur during initial phase of HSV-1 infection in ZF encoded 3-OST-3 receptor. A similar increase in cellular projections was previously reported with other HSV-1 receptors including nectin-1 [35Tiwari V, Oh MJ, Kovacs M, Shukla SY, Valyi-Nagy T, Shukla D. Role for nectin-1 in herpes simplex virus 1 entry and spread in human retinal pigment epithelial cells
FEBS J 2008; 275: 5272-85.].
ZF Encoded 3-OST-3 Mediated HSV-1 Glycoprotein Induced Cell-to-Cell Fusion is Actin Cytoskeleton Dependent
After establishing the role of ZF encoded 3-OST-3 in filopodia induction during HSV-1 entry, we next examined whether ZF 3-OST-3 mediated cell-to-cell fusion is also an actin network dependent process. CHO-K1 cells were used that are resistant to virus-induced cell fusion due to the absence of a gD receptor [28Tiwari V, Clement C, Duncan MB, Chen J, Liu J, Shukla D. A role for 3-O-sulfated heparan sulfate in cell fusion induced by herpes simplex virus type 1
J Gen Virol 2004; 85: 805-9.]. A previously described luciferase reporter gene assay was performed to quantify the induced cell fusion between 3-OS HS cells modified by ZF encoded 3-OST-3 and HSV-1 glycoproteins in presence and absence of Cyto D [28Tiwari V, Clement C, Duncan MB, Chen J, Liu J, Shukla D. A role for 3-O-sulfated heparan sulfate in cell fusion induced by herpes simplex virus type 1
J Gen Virol 2004; 85: 805-9.]. The ‘‘effector’’ CHO-K1 cells were transiently transfected with each of four glycoprotein plasmids: pPEP98 (gB), pPEP99 (gD) pPEP100 (gH), and pPEP101 (gL), as well as the plasmid pT7EMCLuc that expresses a luciferase reporter gene [29Pertel P, Fridberg A, Parish M, Spear PG. Cell fusion induced by herpes simplex virus glycoproteins gB gD and gH-gL requires a gD receptor but not necessarily heparan sulfate
Virology 2001; 279: 313-24.]. The ‘‘target’’ cells were transfected with a 3-OST plasmid expressing ZF encoded 3-OST-3 and the plasmid pCAGT7, which expresses T7 RNA polymerase to induce expression of the luciferase gene. For a negative control, cells were transfected with T7 RNA polymerase and control vector pCDNA3.1. The cells expressing human 3-OST-3 and T7RNA polymerase served as a positive control. As shown in Fig. (4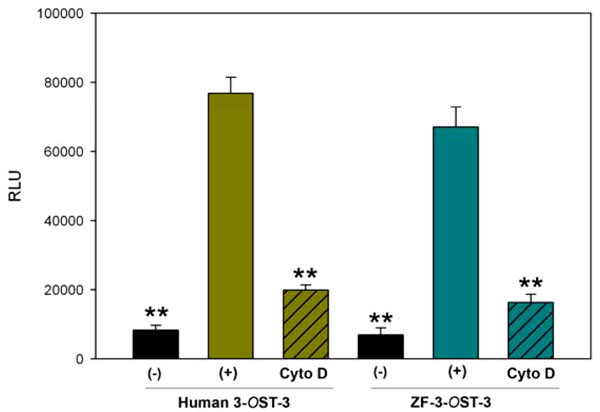 ) a slightly higher amount of fusion occurred in human 3-OST-3 isoform, while ZF encoded 3-OST-3 expressing cells also showed fusion (grey bar) compared to negative control cells without 3-OST-3 expression (black bar). Interestingly significant inhibition in cell-to-cell fusion was observed in presence of Cyto D (grey bars with black lines). These results reinforce our finding that changes in actin cytoskeleton play a critical role during HSV-1 entry and spread and cells expressing ZF 3-OST-3 facilitate filopodia induction.
) a slightly higher amount of fusion occurred in human 3-OST-3 isoform, while ZF encoded 3-OST-3 expressing cells also showed fusion (grey bar) compared to negative control cells without 3-OST-3 expression (black bar). Interestingly significant inhibition in cell-to-cell fusion was observed in presence of Cyto D (grey bars with black lines). These results reinforce our finding that changes in actin cytoskeleton play a critical role during HSV-1 entry and spread and cells expressing ZF 3-OST-3 facilitate filopodia induction.
DISCUSSION
Our previous findings have shown the role of zebrafish (ZF) encoded 3-OST-3 as a receptor for HSV-1 gD [22Hubbard S, Darmani NA, Thrush GR, et al. Zebrafish-encoded 3-O-sulfotransferase-3 isoform mediates herpes simplex virus type 1 entry and spread
Zebrafish 2010; 7: 181-7.]. Here we examined some additional but imoprtant aspects of HSV-1 entry mediated by ZF-3-OST-3 isoform. The novel finding presented herein is that ZF encoded 3-OST-3 receptor cells show significant changes in actin cytoskeleton during initial phase of HSV-1 entry (Fig. 1A , panel c and Fig. 2A
, panel c and Fig. 2A , panel c). Similarly, we demonstrate a role for heparan sulfate, and more importantly modified heparan sulfate, in filopodia induction. Our data suggests a novel role for ZF encoded 3-OST-3 isoform in the induction of filopodia. It has been shown previously that HSV-1 uses filopoida for surfing to efficiently reach the cell body for entry and that surfing is required for enhanced infection [16Oh MJ, Akhtar J, Desai P, Shukla D. A role for heparan sulfate in viral surfing
Biochem Biophys Res Commun 2010; 391: 176-81.]. Although induction of filopodia was not totally dependent on a gD receptor as CHO-K1 cells expressing heparan sulfate (HS) without any 3-OSTs were able to induce filopodia (Fig. 2A
, panel c). Similarly, we demonstrate a role for heparan sulfate, and more importantly modified heparan sulfate, in filopodia induction. Our data suggests a novel role for ZF encoded 3-OST-3 isoform in the induction of filopodia. It has been shown previously that HSV-1 uses filopoida for surfing to efficiently reach the cell body for entry and that surfing is required for enhanced infection [16Oh MJ, Akhtar J, Desai P, Shukla D. A role for heparan sulfate in viral surfing
Biochem Biophys Res Commun 2010; 391: 176-81.]. Although induction of filopodia was not totally dependent on a gD receptor as CHO-K1 cells expressing heparan sulfate (HS) without any 3-OSTs were able to induce filopodia (Fig. 2A , panel b); however, a significant increase in the amount of filopodia was noticed in presence of a gD receptor i.e. CHO-K1 cells expressing either ZF or human form 3-OST-3 receptor. The above finding implies that ZF encoded 3-OST-3 may provide unique gD binding sites for enhanced cell signaling and filopodial development. Specific structures within HS are known to guide retinal axons [36Ogata-Iwao M, Inatani M, Iwao K, et al. Heparan sulfate regulates intraretinal axon pathfinding by retinal ganglion cells
Invest Ophthalmol Vis Sci 2011; 52: 6671-9.]. In addition, HS binding protein, VEGF (vascular endothelial growth factor), is also involved in filopodia formation [37Stenzel D, Lundkvist A, Sauvaget D, et al. Integrin-dependent and -independent functions of astrocytic fibronectin in retinal angiogenesis
Development 2011; 138: 4451-63.]. Therefore, it is likely that actin cytoskeletal changes as a result of HSV-1 gD interaction to ZF-3-OST-3 allow not only virus-cell fusion [22Hubbard S, Darmani NA, Thrush GR, et al. Zebrafish-encoded 3-O-sulfotransferase-3 isoform mediates herpes simplex virus type 1 entry and spread
Zebrafish 2010; 7: 181-7.] but may also enhance viral surfing to enhance viral infectivity.
, panel b); however, a significant increase in the amount of filopodia was noticed in presence of a gD receptor i.e. CHO-K1 cells expressing either ZF or human form 3-OST-3 receptor. The above finding implies that ZF encoded 3-OST-3 may provide unique gD binding sites for enhanced cell signaling and filopodial development. Specific structures within HS are known to guide retinal axons [36Ogata-Iwao M, Inatani M, Iwao K, et al. Heparan sulfate regulates intraretinal axon pathfinding by retinal ganglion cells
Invest Ophthalmol Vis Sci 2011; 52: 6671-9.]. In addition, HS binding protein, VEGF (vascular endothelial growth factor), is also involved in filopodia formation [37Stenzel D, Lundkvist A, Sauvaget D, et al. Integrin-dependent and -independent functions of astrocytic fibronectin in retinal angiogenesis
Development 2011; 138: 4451-63.]. Therefore, it is likely that actin cytoskeletal changes as a result of HSV-1 gD interaction to ZF-3-OST-3 allow not only virus-cell fusion [22Hubbard S, Darmani NA, Thrush GR, et al. Zebrafish-encoded 3-O-sulfotransferase-3 isoform mediates herpes simplex virus type 1 entry and spread
Zebrafish 2010; 7: 181-7.] but may also enhance viral surfing to enhance viral infectivity.
The presented SEM images indicate that pronounced filopodia induction with phagoctyic arms is observed during HSV-1 entry into ZF-3-OST-3 expressing CHO-K1 cells (Fig. 1A , panel c, and Fig. 2A
, panel c, and Fig. 2A , panel c). The filopodia associated arms were similar to the HSV-1 phagoctyic-like uptake by human corneal fibroblasts (CF) cells where 3-OST-3 plays a major role for HSV-1 entry and spread [30Clement C, Tiwari V, Scanlon PM, Vali-Nagy T, Yue BYJT, Shukla D. A novel for phagocytosis-like uptake in herpes simplex virus entry
J Cell Biol 2006; 174: 1009-21.]. These findings support previous studies that HS and 3-OS HS work at two different levels to promote HSV infection. First, it is well documented that HSV-1 glycoprotein B (gB) contact to cell surface HS to induce filopodia which aid in lateral viral movement along the length of filopodia to bring the virions closer to the cell body [16Oh MJ, Akhtar J, Desai P, Shukla D. A role for heparan sulfate in viral surfing
Biochem Biophys Res Commun 2010; 391: 176-81.]. Second, HSV-1 glycoprotein D (gD) interaction to 3-OST-3 receptor triggers the viral uptake via filopodia [30Clement C, Tiwari V, Scanlon PM, Vali-Nagy T, Yue BYJT, Shukla D. A novel for phagocytosis-like uptake in herpes simplex virus entry
J Cell Biol 2006; 174: 1009-21.]. The role of actin cytoskeleton is widely implicated in microbial pathogenesis [38Döhner K, Sodeik B. The role of the cytoskeleton during viral infection
Curr Top Microbiol Immunol 2005; 285: 67-108., 39Greber UF, Way M. A superhighway to virus infection
Cell 2006; 124: 741-54.]. Multiple viruses exploit the host actin cytoskeleton to facilitate important aspects of their lifecycles including viral surfing, entry into target cell, egress, and intercellular spread [40Radtke K, Döhner K, Sodeik B. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell
Cell Microbiol 2006; 3: 387-400.].
, panel c). The filopodia associated arms were similar to the HSV-1 phagoctyic-like uptake by human corneal fibroblasts (CF) cells where 3-OST-3 plays a major role for HSV-1 entry and spread [30Clement C, Tiwari V, Scanlon PM, Vali-Nagy T, Yue BYJT, Shukla D. A novel for phagocytosis-like uptake in herpes simplex virus entry
J Cell Biol 2006; 174: 1009-21.]. These findings support previous studies that HS and 3-OS HS work at two different levels to promote HSV infection. First, it is well documented that HSV-1 glycoprotein B (gB) contact to cell surface HS to induce filopodia which aid in lateral viral movement along the length of filopodia to bring the virions closer to the cell body [16Oh MJ, Akhtar J, Desai P, Shukla D. A role for heparan sulfate in viral surfing
Biochem Biophys Res Commun 2010; 391: 176-81.]. Second, HSV-1 glycoprotein D (gD) interaction to 3-OST-3 receptor triggers the viral uptake via filopodia [30Clement C, Tiwari V, Scanlon PM, Vali-Nagy T, Yue BYJT, Shukla D. A novel for phagocytosis-like uptake in herpes simplex virus entry
J Cell Biol 2006; 174: 1009-21.]. The role of actin cytoskeleton is widely implicated in microbial pathogenesis [38Döhner K, Sodeik B. The role of the cytoskeleton during viral infection
Curr Top Microbiol Immunol 2005; 285: 67-108., 39Greber UF, Way M. A superhighway to virus infection
Cell 2006; 124: 741-54.]. Multiple viruses exploit the host actin cytoskeleton to facilitate important aspects of their lifecycles including viral surfing, entry into target cell, egress, and intercellular spread [40Radtke K, Döhner K, Sodeik B. Viral interactions with the cytoskeleton: a hitchhiker's guide to the cell
Cell Microbiol 2006; 3: 387-400.].
CONCLUSION
The knowledge that HSV-1 entry into ZF 3-OST-3 cells involves filopodial induction can be useful for the following reasons: 1) it can lead the way for the development of anti-ZF-3-OST-3 compounds that can potentially block HSV-1 entry and spread at an early step of viral infection i.e. during induction of filopodia. 2) Because the 3-OST-3 gene family is differentially regulated in ZF [21Cadwallader AB, Yost HJ. Combinatorial expression patterns of heparan sulfate sulfotransferase in zebrafish I The 3-O-sulfotransferase family Dev Dynam 2006; 235: 3423-1.], an important question that remains to be demonstrated is whether ZF encoded 3-OST-3 isoform is unique for filopodia development or other isoforms of ZF encoded 3-OS HS can also function as a gD receptor and allow filopodial induction and 3) A ZF model can be also established to study the in vivo significance of filopodia during HSV-1 infection.
In summary, apart from showing the significance of ZF encoded 3-OST-3 in filopodia induction, our study also implicates HS in filopodia induction, which is sure to pave the way for examining the roles of HS and modified HS in cellular signaling associated with cytoskeletal changes. Future understanding regarding HSV-1 usage of HS and 3-OS HS to facilitate viral entry and spread via filopodia in ZF model is likely to open up new ways to develop anti-HSV agents and strategies to prevent both viral spread and inflammation.
METHODS
Plasmids
The Zebrafish encoded 3-OST-3 gene was cloned into pDream2.1 plasmid vector (Genscript), while the Human 3-OST-3 expressing plasmid (pDS43) was provided by Dr. Shukla (University of Illinois at Chicago) [11Shukla D, Liu J, Blaiklock P, et al. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry Cell 1999; 99: 13-22.]. The HSV-1 (KOS) glycoprotein expressing plasmids used were pPEP98 (gB), pPEP99 (gD), pPEP100 (gH), and pPEP101 (gL) [28Tiwari V, Clement C, Duncan MB, Chen J, Liu J, Shukla D. A role for 3-O-sulfated heparan sulfate in cell fusion induced by herpes simplex virus type 1 J Gen Virol 2004; 85: 805-9.]. Other plasmids used in this study include pCAGT7 (T7 RNA polymerase), and pT7EMCLuc (luciferase gene) for the luciferase assay [29Pertel P, Fridberg A, Parish M, Spear PG. Cell fusion induced by herpes simplex virus glycoproteins gB gD and gH-gL requires a gD receptor but not necessarily heparan sulfate Virology 2001; 279: 313-24.], and a control empty vector pCDNA3.1 from Invitrogen (Carlsbad, CA, USA).
Cell Culture and Viruses
Wild-type Chinese Hamster ovary-K1 (CHO-K1) and heparan sulfate defective cells (CHO-745) were kindly provided by P.G. Spear (Northwestern University, Chicago). Both CHO-K1 and CHO-745 cells were grown in Ham’s F-12 medium (Gibco/BRL, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), and penicillin and streptomycin (Gibco/BRL). The β-galactosidase expressing recombinant HSV-1 (KOS) gL86 was provided by P.G. Spear (Northwestern University, Chicago).
Scanning Electron Microscopy (SEM)
Wild-type CHO-K1, CHO-745 cells and CHO-K1 cells expressing human (H) and zebrafish (ZF) 3-OST-3 receptor were infected with HSV-1 (KOS) at 25 pfu/cell for 45 min at 37oC in triplicate experiments. In a parallel experiment, the CHO-K1 cells expressing ZF-3-OST-3 were pre-incubated with 1× PBS or with an actin polymerizer cyto-D followed by HSV-1 infection at 25 pfu/cell for 45 min at 37oC. The cells were then fixed with 2% formaldehyde/4% glutaraldehyde in 1 × phosphate buffer saline (PBS) prior to SEM study. This was followed by fixing cells with 1% osmium tetroxide formaldehyde/glutaraldehyde for 40 minutes. Dehydration was done using 25% ethanol, 50% ethanol, 70% ethanol, 90% ethanol, 95% ethanol, 100% ethanol at five minutes each respectively. 100% ethanol was repeated to ensure dehydration. Cover slips were removed from dishes and mounted on aluminum studs previously cleaned with 100% ethanol. Cover slip edges were painted with colloidal silver for conduction and dried in a Critical Point Dryer (Samdri-780A). Samples were then coated with gold using a Sputter Coater (Hummer VI-A) for 2.5 minutes. Samples were viewed using a Hitachi S-2700 Scanning Electron Microscope (SEM). Images were captured at 1000-5000x using Revolution image capture system and a controller/analysis system for energy dispersive X-ray spectroscopy.
HSV-1 Entry Assay
As previously described [22Hubbard S, Darmani NA, Thrush GR, et al. Zebrafish-encoded 3-O-sulfotransferase-3 isoform mediates herpes simplex virus type 1 entry and spread Zebrafish 2010; 7: 181-7.] CHO-K1 cells were grown in 6-well plates to subconfluence and transfected with 2.5 μg of human and or ZF encoded 3-OST isoforms (3-OST-3) or control plasmid (pDream2.1 or pCDNA3.1) using LipofectAMINE (Gibco/BRL) [22Hubbard S, Darmani NA, Thrush GR, et al. Zebrafish-encoded 3-O-sulfotransferase-3 isoform mediates herpes simplex virus type 1 entry and spread Zebrafish 2010; 7: 181-7.]. At 16 h post-transfection, the cells were replated into 96-well dishes for pre-treatment with actin depolymerizers (cyto-D and lant-B at indicated concentrations for 45 min at room temperature) followed by infection with β-galactosidase expressing recombinant HSV-1 gL86 virus. After 6-h post infection, β-galactosidase assay were performed using a soluble substrate o-nitrophenyl-β-D-galactopyranoside (ONPG; ImmunoPure, Pierce). The enzymatic activity was measured at 410 nm using a micro-plate reader [22Hubbard S, Darmani NA, Thrush GR, et al. Zebrafish-encoded 3-O-sulfotransferase-3 isoform mediates herpes simplex virus type 1 entry and spread Zebrafish 2010; 7: 181-7.].
HSV-1 Glycoprotein Induced Cell-Fusion Assay
A cell-to-cell fusion assay described previously was used [22Hubbard S, Darmani NA, Thrush GR, et al. Zebrafish-encoded 3-O-sulfotransferase-3 isoform mediates herpes simplex virus type 1 entry and spread Zebrafish 2010; 7: 181-7., 28Tiwari V, Clement C, Duncan MB, Chen J, Liu J, Shukla D. A role for 3-O-sulfated heparan sulfate in cell fusion induced by herpes simplex virus type 1 J Gen Virol 2004; 85: 805-9.]. CHO-K1 cells were grown in 6-well plates to subconfluent levels. The cultured CHO-K1 ‘‘target’’ cells were transfected with plasmids expressing either human or zebrafish (ZF) 3-OST-3 isoform and the luciferase gene. The ‘‘effector’’ or virus-like cells were co-transfected with four HSV-1(KOS) glycoproteins as previously described [28Tiwari V, Clement C, Duncan MB, Chen J, Liu J, Shukla D. A role for 3-O-sulfated heparan sulfate in cell fusion induced by herpes simplex virus type 1 J Gen Virol 2004; 85: 805-9.]. In either case, the total amount of DNA used for transfection was kept constant. For transfection, CHO-K1 cells were grown to 70% confluency in a 6 well dishes. 2.5 µg of plasmid DNA was mixed with 8 µl of Lipofectamine (Gibco/BRL) in to a total volume of 1 ml with serum-free media. After 16 h, target and effector cells were mixed in a 1:1 ratio and then replated in 24-well dishes. In parallel effector cells transfected only with pCDNA3.1 along with T7 RNA polymerase was mixed with the target cells expressing either human or ZF 3-OST-3 isoform was used as a negative control. The activation of the reporter luciferase gene as a measure of cell fusion was examined after 24 h. Luciferase activity was quantified using the luciferase reporter assay system (Promega, Madison, WI).
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
We sincerely thank Dr. Spear (Northwestern University, Chicago) for providing the reagents and the facility at California State University (San Bernardino) for electron microscopy. This work was supported by National Institutes of Health RO1 grants AI057860 (to D. S.) and R15 (1R15AI088429-01A1) to V.T.






