- Home
- About Journals
-
Information for Authors/ReviewersEditorial Policies
Publication Fee
Publication Cycle - Process Flowchart
Online Manuscript Submission and Tracking System
Publishing Ethics and Rectitude
Authorship
Author Benefits
Reviewer Guidelines
Guest Editor Guidelines
Peer Review Workflow
Quick Track Option
Copyediting Services
Bentham Open Membership
Bentham Open Advisory Board
Archiving Policies
Fabricating and Stating False Information
Post Publication Discussions and Corrections
Editorial Management
Advertise With Us
Funding Agencies
Rate List
Kudos
General FAQs
Special Fee Waivers and Discounts
- Contact
- Help
- About Us
- Search

The Open Virology Journal
(Discontinued)
ISSN: 1874-3579 ― Volume 15, 2021
Improving Clinical Laboratory Efficiency: Introduction of Systems for the Diagnosis and Monitoring of HIV Infection
Marta Alvarez, Natalia Chueca, Vicente Guillot, María del Carmen Bernal, Federico García*
Abstract
Since the first tests for identifying individuals with suspected human immunodeficiency virus (HIV) infection were introduced in the mid-1980s, diagnostic virology testing has greatly evolved. The technological advances, automating in the laboratories and the advances in molecular biology techniques have helped introduce invaluable laboratory methods for managing HIV patients. Tests for diagnosis, specially for screening HIV antibodies, are now fully automated; in the same way, tests for monitoring HIV viral load (HIV RNA copies/ml of plasma), which is used for monitoring infection and response to antiretroviral treatment, are also fully automated; however, resistance testing, tropism determination and minor variant detection, which are used to make decisions for changing antiretroviral treatment regimens in patients failing therapy, still remain highly laborious and time consuming. This chapter will review the main aspects relating to the automating of the methods available for laboratory diagnosis as well as for monitoring of the HIV infection and determination of resistance to antiretrovirals and viral tropism.
Article Information
Identifiers and Pagination:
Year: 2012Volume: 6
Issue: Suppl 1
First Page: 135
Last Page: 143
Publisher Id: TOVJ-6-135
DOI: 10.2174/1874357901206010135
Article History:
Received Date: 15/7/2012Revision Received Date: 3/8/2012
Acceptance Date: 6/8/2012
Electronic publication date: 30/11/2012
Collection year: 2012
open-access license: This is an open access article licensed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted, non-commercial use, distribution and reproduction in any medium, provided the work is properly cited.
* Address correspondence to this author at the Servicio de Microbiología, Hospital Universitario San Cecilio, Av, Dr Oloriz 16, Granada, Spain; Tel: 0034-958023865; E-mail: fegarcia@ugr.es
| Open Peer Review Details | |||
|---|---|---|---|
| Manuscript submitted on 15-7-2012 |
Original Manuscript | Improving Clinical Laboratory Efficiency: Introduction of Systems for the Diagnosis and Monitoring of HIV Infection | |
INTRODUCTION
Since the first tests for identifying individuals with suspected human immunodeficiency virus (HIV) infection were introduced in the mid-1980s, diagnostic virology testing has greatly evolved thanks to increased understanding of immunopathogenic mechanisms, the host-virus relationship, viral replication mechanisms and the immune response that occurs in infected people over the course of the infection.
The technological advances, automating in the laboratories and the advances in molecular biology techniques have helped introduce invaluable laboratory methods for managing HIV patients. During the course of HIV infection, various viral markers can be used to identify the infection and monitor the treatment. Each has different kinetics and appears at a different time and so the choice of marker to be detected will depend on the objective of the diagnosis [1Buttó S, Suligoi B, Fanales-Belasio E, Raimondo M. Laboratory diagnostics for HIV infection Ann Ist Super Sanitá 2010; 46(1 ): 24-33.]. The first marker to appear after infection is HIV RNA, which can be detected by amplification techniques around 2 weeks after infection, generally at 10-12 days. At more or less the same time as the HIV RNA, HIV DNA integrated into the cell genome (proviral DNA) can be detected. The p24 antigen appears in serum at 11-13 days and can be detected using maximum-sensitivity techniques for about one and a half months [2Tebourski F, Slim A, Elgaaied A . The significance of combining World Health Organization and Center for Disease Control criteria to resolve indeterminate human immunodeficiency virus tipe-1 Western blot results Diagn Microbiol Infect Dis 2004; 48: 59-61.]. Antibodies can be detected in serum at 3-4 weeks after infection (average 22 days), reaching peak concentration at 10-12 weeks [3Fanales-Belasio E, Raimondo M, Suligoi B, Butto S. HIV virology and pathogenetic mechanisms of infection: a brief overview Ann Ist Super Sanit 2010; 46(1 ): 5-14.]. When the antibodies appear, the viraemia levels diminish and the p24 antigen disappears as a result of the formation of immunocomplexes [1Buttó S, Suligoi B, Fanales-Belasio E, Raimondo M. Laboratory diagnostics for HIV infection Ann Ist Super Sanitá 2010; 46(1 ): 24-33.]. The interval between infection and the appearance of antibodies or seroconversion is known as the window period and is characterised by the presence of HIV RNA, proviral DNA, and p24 antigen and the absence of specific antibodies.
Laboratory diagnosis of HIV infection is mainly done through demonstration of the presence of anti-HIV antibodies, for which screening and confirmation techniques have to be used [4García F, Álvarez M, Bernal C, Chueca N, Guillot V. Laboratory diagnosis of HIV infection, viral tropism and resistance to antiretrovirals Enferm Infect Microbiol Clin 2011; 29(4 ): 297-307.]. Determination of proviral DNA is used at times, primarily for diagnosis of vertically-transmitted infection. Once HIV infection is confirmed, HIV viral load (HIV RNA expressed in copies/ml of plasma) and resistance testing are used to evaluate the efficacy of antiretroviral therapy, and to design new treatment regimens in patients with treatment failure. A new tool to help design the new treatment regimen is the determination of viral tropism.
This chapter will review the main aspects relating to the automating of the methods available for laboratory diagnosis as well as for monitoring of the HIV infection and determination of resistance to antiretrovirals and viral tropism.
DIAGNOSIS OF HIV INFECTION
HIV Infection Detection Techniques
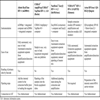
Infections are diagnosed by detecting the presence of specific antibodies, since they are found in the serum in virtually 100% of infected people. With the aim of minimising the risk of false-negative results, all the techniques are highly sensitive, with close to 99% accuracy. It is difficult to achieve 100% sensitivity as seroconversion does not take place until 3-4 weeks after infection and, moreover, infected people can be seronegative as a result of immunity defects. The increase in sensitivity entails a reduction in specificity, which with the current techniques is around 99%. Furthermore, a lower prevalence of HIV infection in the studied population reduces the positive predictive value, meaning that the likelihood of false-positive results occurring with low HIV infection rates is higher. All positive results must therefore be confirmed by a confirmatory test [1Buttó S, Suligoi B, Fanales-Belasio E, Raimondo M. Laboratory diagnostics for HIV infection Ann Ist Super Sanitá 2010; 46(1 ): 24-33.].
Screening Techniques
ELISA.
The first techniques were introduced in diagnosis in 1985 but they have greatly evolved since then. Now, third-generation ELISA are used with “sandwich” format which detect IgG and IgM antibodies and antibodies to all the M subtypes, groups N and O and HIV-2. Fourth-generation techniques have recently been introduced which allow simultaneous detection of antibodies and p24 antigen, reducing the window period to 13-15 days [5Weber B. Screening of HIV infection:role of molecular and immunological assays Expert Rev Mol Diagn 2006; 6: 399-411.]. With these techniques, the sensitivity is increased to 99.9%. This reduces the likelihood of a false-negative result and means that in principle, a negative result does not require either confirmation or serological monitoring except in people at high risk of acquiring the infection [6Butto S, Raimondo M, Fanales-Belasio E, Suligoi B. Suggest strategies for the laboratory Diagnosis of HIV infection in Italy Ann IST Super Sanita 2010; 46(1 ): 34-41.]. The specificity is between 99.5% and 99.9%. However, false positives can occur as a result of non-specific recognition of substances in the serum by the virus antigens from the antigen base.
ELFA
(enzyme linked immunofluorescent assay). These tests are modified versions of the ELISA techniques that use solid phases with a larger contact surface area, thereby reducing incubation times. As label, they use an enzyme and fluorescent substrates which are transformed into a fluorescent product by the action of the enzyme. The rates of fluorescence emitted are measured with a fluorescence detector. These techniques enable the process to be automated, the processing of a large number of samples, less handling of the sample (which reduces the number of errors), a reduction in costs and, most importantly, a reduction in the duration of the process, with first result response times of less than 60 minutes [7Nazida F. Laboratory test for detection of human Inmunodeficiency virus type 1 infection Clin Diagn Lab Inmunol 1995; 2: 637-45.]. Third- and fourth-generation techniques are both currently used.
Chemiluminescence.
These techniques use chemiluminescent compounds to label the antigens or antibodies which, when put in contact with a substrate and an oxidant, form an unstable intermediate compound. When this returns to its ground state, it becomes excited and emits energy in the form of light which is measured by a photomultiplier tube. Various chemiluminescent compounds can be used, such as luminol, dioxetane, ruthenium and acridine. These techniques have been adapted to large analysers and have many advantages as: they are totally automated; primary tubes with barcode can be used; reagents can be kept refrigerated; long stability of the calibration curve; 200-400 tests/hour; response time of 30-60 minutes; they make it possible to design conditional test methods; and bidirectional computer connection is possible. Third- and fourth-generation methods are currently available.
Confirmatory Tests
The most commonly-used confirmatory techniques are Western Blot (WB) and recombinant immunoblot or line immunoassay (LIA). They have at least the same sensitivity as ELISA and superior specificity. Both techniques can incorporate HIV-2 envelope antigens, thereby making it possible to diagnose HIV-2.
The methodology for both methods has now been semi-automated, making them easier to perform. However, the results can be subjective as the reading is based on observation of the presence of coloured bands which correspond to different viral proteins [7Nazida F. Laboratory test for detection of human Inmunodeficiency virus type 1 infection Clin Diagn Lab Inmunol 1995; 2: 637-45.]. Each laboratory must therefore establish a strict system for taking reactivity readings.
Detection of Antigenaemia
The detection of specific antibodies indicates exposure to the virus and infection and direct detection of the viral p24 antigen introduces a dynamic concept to serology; since it is a marker of virus replication, it provides information on the current status of the infection, it is detected in initial stages of the infection or in the development of AIDS, and it serves as back-up to the serological diagnosis in situations where the detection of antibodies is inconclusive [8Casey JM, Kim Y, Andersen PR. Human T-cell lymphotropic virus type III: immunologic characterization and primary structure analysis of the major internal protein p24 J Virol 1985; 55: 417-2.]. The p24 antigen can be measured in serum and plasma with uptake ELISA techniques that increase sensitivity, which at present can be as high as 8 pg/ml.
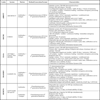
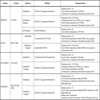
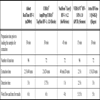
The main characteristics of the available systems for screening and confirmation of HIV infection and the extent to which they are automated are shown below. Table 1 shows the methodology, generation, format and main characteristics of fully automated systems for HIV diagnosis. Table 2 lists the main features of ELISA semi-automated systems. Table 3 shows the confirmatory assays and their features.
VIROLOGICAL MARKERS FOR MONITORING HIV INFECTION
HIV Viral Load
Viral load, or plasma viraemia, is the amount of virus circulating in plasma and refers to the number of copies of HIV RNA present per millilitre, expressed as copies/ml or log10. The HIV viral load and CD4 lymphocyte count are currently the two most used prognostic markers of clinical progression of the HIV infection [9Korenromp EL, Williams BG, Schmid GP, Dye C. Clinical prognostic value of RNA viral load and CD4 cell counts during untreated HIV-1 infection--a quantitative review PLoS One 2009; 17(4(6) ): e5950.]; the first is an indicator of virus replication and the second reflects how the host’s immune system is working. Both parameters are monitored every 3-6 months, although some authors [10Panel on Antiretroviral Guidelines for Adults and Adolescents Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents Department of Health and Human Services 2012 March; Available at http://www.aidsinfo.nih.gov/contentfiles/ lvguidelines/adultandadolescentgl.pdf] are now advocating annual monitoring, at least of the CD4 count. A high initial viral load is a marker of rapid progression and clinical progression is always preceded by an increase in the viral load. Changes in plasma HIV-1 RNA levels are the best marker for response to antiretroviral therapy, since they are sensitive, fast and reliable in terms of good management of the patient with HIV infection. The success of antiretroviral therapy is defined in the majority of guidelines as the suppression of plasma viraemia and clinically, it is considered as such when the patient has two successive viral loads below 50 copies/ml [11Hirsch MS, Günthard HF, Schapiro JM, et al. International AIDS Society-USA. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel Top HIV Med 2008; 16(3 ): 266-85.], although this cut-off point has been the subject of debate in the last year [12Doyle T, Smith C, Vitiello P, et al. Plasma HIV-1 RNA detection below 50 copies/ml and risk of virologic rebound in patients receiving highly active antiretroviral therapy Clin Infect Dis 2012; 54(5 ): 724-32.-14Alvarez M, Chueca N, Guillot V, Peña A, Muñoz L, Parra J. Plasma HIV-1 viral load below 50 using the Cobas AmpliPrep Cobas TaqMan HIV-1 test Roche is aldo associated to a higher risk of virologic rebound In: 10th European Meeting on HIV & Hepatitis Treatment Strategies & Antiviral Drug Resistance; Barcelona, Spain; 28-30 March 2012. Abstract O36].
Viral load should not be used routinely to diagnose the infection, except in special cases such as confirmation of neonatal infection.
Given how important it is to measure this parameter, accurate, reproducible, cost-effective techniques are required [15Luft LM, Gill MJ, Church DL. HIV-1 viral diversity and its implications for viral load testing: review of current platforms Int J Infect Dis 2011; 15(10 ): e661-70.-17Cobb BR, Vaks JE, Do T, Vilchez RA. Evolution in the sensitivity of quantitative HIV-1 viral load tests J Clin Virol 2011; 52S: S77-82.].
There are currently a number of different platforms available for quantifying HIV viral load:
- Abbott RealTime HIV-1 m2000rt (Abbott Molecular Diagnostics): combines the extraction in the automated m2000sp system and qrtPCR amplification and detection with the automated m2000rt equipment.
- COBAS AmpliPrep TaqMan HIV-1 48 (Roche Molecular Diagnostics): uses the COBAS AmpliPrep for the extraction of the sample and the COBAS TaqMan 48 v2.0 for the qrtPCR amplification and detection.
- NucliSens EasyQ HIV-1 v1.2 (bioMérieux): uses the EasyMAG for the extraction and the EasyQ for qrtPCR amplification and detection.
- VERSANT HIV-1 RNA 1.0 (kPCR) (Siemens): this system combines a sample extraction module (SP module) with an amplification and detection module (AD module).
- Artus HIVirus-1 QIAsymphony SP/AS and Rotor-Gene Q (Qiagen): the extraction is carried out in the QIAsymphonySP/AS equipment and the amplification and detection in the Rotor-Gene Q.
All of these use real-time PCR to quantify HIV RNA in plasma, displacing previously-used techniques such as conventional PCR, the NASBA technology, ligase chain reaction or signal amplification. The advantages of real-time PCR are its high levels of analytical sensitivity, reproducibility and linearity, dynamic range and the fact that the current techniques can quantify different types and subtypes of HIV, not only HIV-1 subtype B. These instruments incorporate automated RNA extraction methods which significantly reduce the time it takes to obtain results and the risk of contamination, and a larger number of samples can be processed. They all start with the reverse transcription of HIV-1 RNA to cDNA. The target is then amplified with real-time PCR, generating fluorescent molecules which are ligated to oligonucleotide probes which specifically bind to the amplified product. When the fluorescence exceeds a minimum signal, the number of PCR cycles (Ct) that have taken place is used for the quantification. One of the advantages of this technology is that it enables detection and quantification of the target without having to handle the reaction tubes. The main features of these assays are shown in Tables 4-7.
There is intrinsic variability between the different assays and it is therefore recommended that the monitoring of a patient’s viral load is always done with the same system; if a change of assay is unavoidable, it must be ensured that a new baseline quantification of the HIV-1 RNA is obtained with the technique to be used thereafter. For quantification in the different subtypes of HIV, nowadays, all these techniques have been adapted to detect a large number of non-B subtypes.
Resistance Testing
Drug resistance testing has become a key component of proper HIV clinical care and is currently recommended by most HIV treatment guidelines [11Hirsch MS, Günthard HF, Schapiro JM, et al. International AIDS Society-USA. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel Top HIV Med 2008; 16(3 ): 266-85., 18European AIDS Clinical Society (EACS) guidelines for the clinical management and treatment of HIV-infected adults. Clumeck N, Pozniak A, Raffi F; EACS Executive Committee HIV Med 2008; 9(2 ): 65-71., 19National consensus document by GESIDA/National Aids Plan on antiretroviral treatment in adults infected by the human immunodeficiency virus (January 2011 update). Panel de expertos de GESIDA y Plan Nacional sobre el Sida. Enferm Infecc Microbiol Clin 2011; 29(3 ): 209.e1-103.] as the standard of care, both in terms of choosing the most effective antiretroviral therapy for first-line regimens, by investigating the transmission of drug-resistant variants at the time of diagnoses (primary resistance) and in terms of the selection of active drugs in subsequent failures to first- or further-treatment-line failures (acquired resistance).
Although resistance assays can be categorized as either phenotyping or genotyping, the genotyping assays are the most widely-adopted in routine diagnostic laboratories. Phenotyping resistance assays measure the extent to which an antiretroviral drug inhibits virus replication in vitro and is usually performed by demonstrating a change in the inhibitory concentration (IC) that is required to inhibit in vitro growth by 50 percent (IC50) compared with virus replication in the absence of drug. Results are reported as a fold change in drug susceptibility of the patient sample in relation to a wild type reference strain. These assays are currently performed using Recombinant Virus Assay technology. They are typically available from commercial laboratories and are rarely performed in routine diagnostic laboratories. An overview of the phenotyping methods and their advantages and limitations can be found in Garcia et al. [4García F, Álvarez M, Bernal C, Chueca N, Guillot V. Laboratory diagnosis of HIV infection, viral tropism and resistance to antiretrovirals Enferm Infect Microbiol Clin 2011; 29(4 ): 297-307.].
Genotyping resistance assays detect the presence of specific drug-resistance mutations. Mutations are then interpreted into a drug-resistance report by using resistance interpretation systems. The report typically gives a classification into “susceptible", "possibly resistant" or "resistant" for each antiretroviral agent. Interpretation rules and algorithms can vary greatly from one to another [20García F, Palomares JC, Martínez NM, et al. Alvarez Study of different systems for interpreting results of genotypic antiretroviral drug resistance tests Antivir Ther 2003; 8: 251-52.]. Currently the protease (Pro), reverse transcriptase (RT), integrase (INT) and the V3 loop are the regions of interest for HIV-1 DNA sequencing. Pro, RT and INT are used to investigate resistance to protease, reverse transcriptase (nucleoside/otide and non-nucleoside analogues) and integrase inhibitors, while V3 sequencing is used for co-receptor usage estimation (tropism testing) prior to the use of CCR5 antagonists.
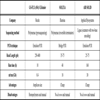
The most widely-used tests in routine clinical laboratories are those investigating RT and Pro mutations. Two commercially available FDA-approved and CE-marked kits are available: TRUGENE HIV-1 Genotyping Assay (Siemens NAD) and ViroSeq® HIV-1 Genotyping System (Celera-Abbott diagnostics). Although there are some differences in performance, both tests are labour intensive, both for the sequence generation and detection steps. Both methods generally provide concordant results in the way they detect mutations, but they differ in the way resistance reports are given [21Kozal M. Overview of HIV drug resistance testing assays Avaiable from: http://www.uptodate.com/ ].
Compared to phenotyping, the advantages of genotyping tests include less complexity, shorter turn-around time and lower cost, while their main limitations are: being an indirect measure of resistance; that they only interrogate known mutations; the difficulty in interpretation when complex patterns of mutations are present; the differential effect on resistance that combinations of individual mutations may have compared to individual mutations alone; and that they cannot detect low-abundance resistant mutations (minor variants).
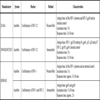
Table 8 shows the main features of two tests, including technical, complexity, biosafety and cost-effectiveness aspects.
Minority Variant Detection and Tropism Testing
Conventional resistance tests can detect mutations which comprise over 20% of all the viral genomes amplified with RT-PCR. We are now at the stage where it is becoming important to detect mutations that might be between 1% and 20%, since it has been demonstrated that these patients are at greatest risk of failing therapy [22Li JZ, Paredes R, Ribaudo HJ, et al. Low-Frequency HIV-1 Drug Resistance Mutations and Risk of NNRTI-Based Antiretroviral Treatment Failure: A Systematic Review and Pooled Analysis JAMA 2011; 305(13 ): 1327-35.]. Techniques are therefore being introduced which are capable of detecting and/or quantifying mutation levels below the 20% threshold imposed by the currently-available techniques.
One of the methods already available, allele-specific real-time PCR (AS-PCR), can detect mutations present in as low as 0.01% of the population as a whole [23Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naïve populations and associate with reduced treatment efficacy PLoS Med 2008; 5(7 ): e158.]. Despite this extraordinary sensitivity, however, the great disadvantage of this method is that an assay for every allele of every mutation is needed. This has meant that the only results presented have been for detecting the main mutations associated with resistance to non-nucleoside analogues (K103N, Y181C, G190A) and M184V [23Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naïve populations and associate with reduced treatment efficacy PLoS Med 2008; 5(7 ): e158.-25Geretti AM, Fox ZV, Booth CL, et al. Low-frequency K103N strengthens the impact of transmitted drug resistance on virologic responses to first-line efavirenz or nevirapine-based highly active antiretroviral therapy J Acquir Immune Defic Syndr 2009; 52(5 ): 569-73.], a mutation that has been associated with resistance to the main nucleoside analogues.
This drawback is overcome by the massively ultra deep parallel sequencing techniques (UDPS) which have been developed in the last few years and are able to detect mutations from 0.5%. Moreover, as these are single genome sequencing techniques, they can provide data on the percentage of each mutation compared to the whole population and on the mutational load, which is proving to be extremely useful as a true parameter in the interpretation of resistances. Another advantage of these platforms is that by sequencing single genomes, we are not limited to the known mutations, but with the introduction of the new antiretrovirals, we will be able to read each of the potential positions of interest in the future.
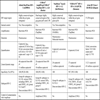
There are currently various UDPS platforms available. Table 9 provides a summary of the different characteristics of the platforms which are more advanced in terms of development, highlighting two principal aspects. The first is the length of the readings generated which, with the Roche platform (454) [26Schuster SC. Next-generation sequencing transforms today's biology Nat Methods 2008; 5(1 ): 16-8., 27Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing Nature 2008; 452(7189 ): 872-6.], is close to that obtained by the Sanger method, making them ideal for single genome sequencing, although the great disadvantage is that they provide poor resolution in homopolymeric sequences. In contrast, Solexa [28Mardis ER. Next-generation DNA sequencing methods Annu Rev Genomics Hum Genet 2008; 9: 387-402.] and SOLiD [29Valouev A, Ichikawa J, Tonthat T, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning Genome Res 2008; 18(7 ): 1051-63.] have the great advantage over pyrosequencing of providing good resolution of the homopolymeric regions, but are not capable of generating readings beyond 75 bases and they cannot be used for de novo sequencing. Another important aspect is that they are able to sequence the DNA without it first having to be cloned, saving a lot of work and laboratory space. With these new sequencers, the cost of each nucleotide has fallen from $10 in 1990 to $0.01 in 2012.
The great disadvantage of these techniques is that they are not automated. Although systems are being developed to enable the different steps to be automated, in the meantime, it has to be said that they are very laborious manual techniques, they take up a lot of time and require highly qualified personnel. So far, these platforms are not being routinely used in the laboratory. However, we are sure that in the not-too-distant future, that will change and these ultra-sequencing techniques will be available for more general use.
With regard to tropism testing, tropism can be inferred using phenotyping methods (Antivirogram® (Virco) [30Van Baelen K, Vandenbrouke I, Rondelez E, Van Eygen V, Vermeiren H, Stuyver L. HIV-1 coreceptor usage determination in clinical isolates using clonal and population-based genotypic and phenotypic assays J Virol Methods 2007; 146: 61-73.], Pheno Sense® (Virologic) [31Coakley E, Petropoulos CJ, Whitcomb JM. Assessing chemokine co-receptor usage in HIV Curr Opin Infect Dis 2005; 18(1 ): 9-15.], Phenoscript® (VIRAlliance) [32Poveda E, Rodés B, Labernardière JL, et al. Evolution of genotypic and phenotypic resistance to Enfuvirtide in HIV-infected patients experiencing prolonged virologic failure J Med Virol 2004; 74: 21-8.] and MT-2 assay [33Coakley E, Reeves JD, Huang W, et al. Comparison of human immunodeficiency virus type 1 tropism profiles in clinical samples by the Trofile and MT-2 assays Antimicrob Agents Chemother 2009; 53(11 ): 4686-93.]), which are only possible in specialised laboratories or private companies that provide the logistics for collection and transport of samples. It can also be determined by using a genotyping method consisting of sequencing the V3 region of the gp140 envelope gene; depending on the sequence the quasi HIV species present, the virus will have R5 tropism (use the co-receptor CCR5) or X4 tropism (use the co-receptor CXCR4). The interest in this determination is that, before being able to use a CCR5 antagonist (the only one approved so far is Maraviroc) it has to be confirmed that the patients are carriers of virus with R5 tropism. At present, there are no commercially-available tests with CE marking or FDA approval for determining viral tropism. These techniques have not been automated in the laboratory and manual procedures have to be used in order to obtain results [34Chueca N, Garrido C, Alvarez M, et al. Improvement in the determination of HIV-1 tropism using the V3 gene sequence and a combination of bioinformatic tools J Med Virol 2009; 81: 763-.].
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
ACKNOWLEDGEMENTS
Declared none.
REFERENCES
| [1] | Buttó S, Suligoi B, Fanales-Belasio E, Raimondo M. Laboratory diagnostics for HIV infection Ann Ist Super Sanitá 2010; 46(1 ): 24-33. |
| [2] | Tebourski F, Slim A, Elgaaied A . The significance of combining World Health Organization and Center for Disease Control criteria to resolve indeterminate human immunodeficiency virus tipe-1 Western blot results Diagn Microbiol Infect Dis 2004; 48: 59-61. |
| [3] | Fanales-Belasio E, Raimondo M, Suligoi B, Butto S. HIV virology and pathogenetic mechanisms of infection: a brief overview Ann Ist Super Sanit 2010; 46(1 ): 5-14. |
| [4] | García F, Álvarez M, Bernal C, Chueca N, Guillot V. Laboratory diagnosis of HIV infection, viral tropism and resistance to antiretrovirals Enferm Infect Microbiol Clin 2011; 29(4 ): 297-307. |
| [5] | Weber B. Screening of HIV infection:role of molecular and immunological assays Expert Rev Mol Diagn 2006; 6: 399-411. |
| [6] | Butto S, Raimondo M, Fanales-Belasio E, Suligoi B. Suggest strategies for the laboratory Diagnosis of HIV infection in Italy Ann IST Super Sanita 2010; 46(1 ): 34-41. |
| [7] | Nazida F. Laboratory test for detection of human Inmunodeficiency virus type 1 infection Clin Diagn Lab Inmunol 1995; 2: 637-45. |
| [8] | Casey JM, Kim Y, Andersen PR. Human T-cell lymphotropic virus type III: immunologic characterization and primary structure analysis of the major internal protein p24 J Virol 1985; 55: 417-2. |
| [9] | Korenromp EL, Williams BG, Schmid GP, Dye C. Clinical prognostic value of RNA viral load and CD4 cell counts during untreated HIV-1 infection--a quantitative review PLoS One 2009; 17(4(6) ): e5950. |
| [10] | Panel on Antiretroviral Guidelines for Adults and Adolescents Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents Department of Health and Human Services 2012 March; Available at http://www.aidsinfo.nih.gov/contentfiles/ lvguidelines/adultandadolescentgl.pdf |
| [11] | Hirsch MS, Günthard HF, Schapiro JM, et al. International AIDS Society-USA. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel Top HIV Med 2008; 16(3 ): 266-85. |
| [12] | Doyle T, Smith C, Vitiello P, et al. Plasma HIV-1 RNA detection below 50 copies/ml and risk of virologic rebound in patients receiving highly active antiretroviral therapy Clin Infect Dis 2012; 54(5 ): 724-32. |
| [13] | Maggiolo F, Callegaro A, Cologni G, et al. Ultrasensitive assessment of residual low-level HIV viremia in HAART-treated patients and risk of virological failure J Acquir Immune Defic Syndr 2012; 60(5 ): 473-82. |
| [14] | Alvarez M, Chueca N, Guillot V, Peña A, Muñoz L, Parra J. Plasma HIV-1 viral load below 50 using the Cobas AmpliPrep Cobas TaqMan HIV-1 test Roche is aldo associated to a higher risk of virologic rebound In: 10th European Meeting on HIV & Hepatitis Treatment Strategies & Antiviral Drug Resistance; Barcelona, Spain; 28-30 March 2012. Abstract O36 |
| [15] | Luft LM, Gill MJ, Church DL. HIV-1 viral diversity and its implications for viral load testing: review of current platforms Int J Infect Dis 2011; 15(10 ): e661-70. |
| [16] | Church D, Gregson D, Lloyd T, et al. Comparison of the RealTime HIV-1, COBAS TaqMan 48 v1.0, Easy Q v1.2, and Versant v3.0 assays for determination of HIV-1 viral loads in a cohort of Canadian patients with diverse HIV subtype infections J Clin Microiol 2011; 49(1 ): 118-24. |
| [17] | Cobb BR, Vaks JE, Do T, Vilchez RA. Evolution in the sensitivity of quantitative HIV-1 viral load tests J Clin Virol 2011; 52S: S77-82. |
| [18] | European AIDS Clinical Society (EACS) guidelines for the clinical management and treatment of HIV-infected adults. Clumeck N, Pozniak A, Raffi F; EACS Executive Committee HIV Med 2008; 9(2 ): 65-71. |
| [19] | National consensus document by GESIDA/National Aids Plan on antiretroviral treatment in adults infected by the human immunodeficiency virus (January 2011 update). Panel de expertos de GESIDA y Plan Nacional sobre el Sida. Enferm Infecc Microbiol Clin 2011; 29(3 ): 209.e1-103. |
| [20] | García F, Palomares JC, Martínez NM, et al. Alvarez Study of different systems for interpreting results of genotypic antiretroviral drug resistance tests Antivir Ther 2003; 8: 251-52. |
| [21] | Kozal M. Overview of HIV drug resistance testing assays Avaiable from: http://www.uptodate.com/ |
| [22] | Li JZ, Paredes R, Ribaudo HJ, et al. Low-Frequency HIV-1 Drug Resistance Mutations and Risk of NNRTI-Based Antiretroviral Treatment Failure: A Systematic Review and Pooled Analysis JAMA 2011; 305(13 ): 1327-35. |
| [23] | Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naïve populations and associate with reduced treatment efficacy PLoS Med 2008; 5(7 ): e158. |
| [24] | Metzner KJ, Giulieri SG, Knoepfel SA, Rauch P, Burgisser P, Yerly S, et al. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients Clin Infect Dis 2009; 48(2 ): 239-47. |
| [25] | Geretti AM, Fox ZV, Booth CL, et al. Low-frequency K103N strengthens the impact of transmitted drug resistance on virologic responses to first-line efavirenz or nevirapine-based highly active antiretroviral therapy J Acquir Immune Defic Syndr 2009; 52(5 ): 569-73. |
| [26] | Schuster SC. Next-generation sequencing transforms today's biology Nat Methods 2008; 5(1 ): 16-8. |
| [27] | Wheeler DA, Srinivasan M, Egholm M, et al. The complete genome of an individual by massively parallel DNA sequencing Nature 2008; 452(7189 ): 872-6. |
| [28] | Mardis ER. Next-generation DNA sequencing methods Annu Rev Genomics Hum Genet 2008; 9: 387-402. |
| [29] | Valouev A, Ichikawa J, Tonthat T, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning Genome Res 2008; 18(7 ): 1051-63. |
| [30] | Van Baelen K, Vandenbrouke I, Rondelez E, Van Eygen V, Vermeiren H, Stuyver L. HIV-1 coreceptor usage determination in clinical isolates using clonal and population-based genotypic and phenotypic assays J Virol Methods 2007; 146: 61-73. |
| [31] | Coakley E, Petropoulos CJ, Whitcomb JM. Assessing chemokine co-receptor usage in HIV Curr Opin Infect Dis 2005; 18(1 ): 9-15. |
| [32] | Poveda E, Rodés B, Labernardière JL, et al. Evolution of genotypic and phenotypic resistance to Enfuvirtide in HIV-infected patients experiencing prolonged virologic failure J Med Virol 2004; 74: 21-8. |
| [33] | Coakley E, Reeves JD, Huang W, et al. Comparison of human immunodeficiency virus type 1 tropism profiles in clinical samples by the Trofile and MT-2 assays Antimicrob Agents Chemother 2009; 53(11 ): 4686-93. |
| [34] | Chueca N, Garrido C, Alvarez M, et al. Improvement in the determination of HIV-1 tropism using the V3 gene sequence and a combination of bioinformatic tools J Med Virol 2009; 81: 763-. |